Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set02_ATM_minus_versus_BT_ATM_minus |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_pos |
| GeneSet | TMTCGCGANR_UNKNOWN |
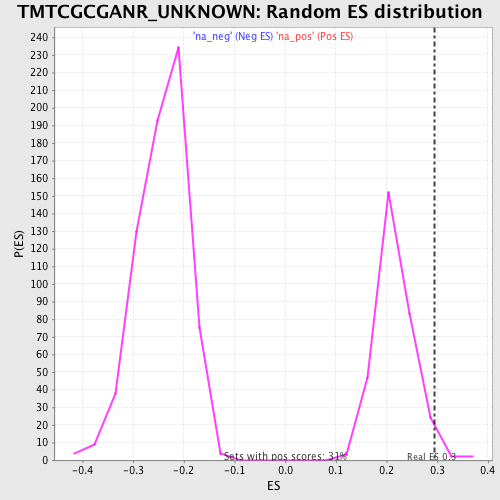
| Enrichment Score (ES) | 0.29487154 |
| Normalized Enrichment Score (NES) | 1.365347 |
| Nominal p-value | 0.028753994 |
| FDR q-value | 0.22991827 |
| FWER p-Value | 0.997 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | MATR3 | 55 | 3.775 | 0.0346 | Yes | ||
| 2 | SLC12A2 | 297 | 2.620 | 0.0493 | Yes | ||
| 3 | ATF2 | 444 | 2.361 | 0.0659 | Yes | ||
| 4 | MAP3K7 | 448 | 2.353 | 0.0888 | Yes | ||
| 5 | ZFP91 | 657 | 2.099 | 0.1000 | Yes | ||
| 6 | PURA | 828 | 1.932 | 0.1112 | Yes | ||
| 7 | TTC14 | 879 | 1.900 | 0.1276 | Yes | ||
| 8 | SEC63 | 1139 | 1.737 | 0.1328 | Yes | ||
| 9 | CD2AP | 1233 | 1.689 | 0.1452 | Yes | ||
| 10 | PIGN | 1480 | 1.570 | 0.1493 | Yes | ||
| 11 | HSPA4 | 1670 | 1.494 | 0.1554 | Yes | ||
| 12 | HNRPK | 1685 | 1.489 | 0.1694 | Yes | ||
| 13 | LUC7L2 | 1778 | 1.447 | 0.1794 | Yes | ||
| 14 | FUS | 1896 | 1.396 | 0.1878 | Yes | ||
| 15 | TLOC1 | 1901 | 1.391 | 0.2012 | Yes | ||
| 16 | PLOD2 | 2000 | 1.360 | 0.2101 | Yes | ||
| 17 | STXBP4 | 2217 | 1.289 | 0.2129 | Yes | ||
| 18 | ASH2L | 2340 | 1.246 | 0.2196 | Yes | ||
| 19 | UHRF2 | 2357 | 1.239 | 0.2310 | Yes | ||
| 20 | DENR | 2712 | 1.149 | 0.2261 | Yes | ||
| 21 | SCAMP5 | 2826 | 1.122 | 0.2320 | Yes | ||
| 22 | UPF3B | 2881 | 1.110 | 0.2404 | Yes | ||
| 23 | PTPN4 | 3009 | 1.081 | 0.2452 | Yes | ||
| 24 | CAMK2D | 3046 | 1.072 | 0.2541 | Yes | ||
| 25 | TLK2 | 3164 | 1.046 | 0.2591 | Yes | ||
| 26 | FGFR1OP2 | 3171 | 1.043 | 0.2690 | Yes | ||
| 27 | PSMB2 | 3343 | 1.003 | 0.2711 | Yes | ||
| 28 | PPP1R12A | 3345 | 1.003 | 0.2809 | Yes | ||
| 29 | SMARCD2 | 3423 | 0.987 | 0.2871 | Yes | ||
| 30 | OFD1 | 3508 | 0.966 | 0.2927 | Yes | ||
| 31 | TROAP | 3663 | 0.936 | 0.2949 | Yes | ||
| 32 | RBPSUH | 4891 | 0.726 | 0.2459 | No | ||
| 33 | UBE2D3 | 4979 | 0.711 | 0.2489 | No | ||
| 34 | TAS1R1 | 4983 | 0.710 | 0.2557 | No | ||
| 35 | DDX3X | 5029 | 0.703 | 0.2606 | No | ||
| 36 | MOBKL1A | 5130 | 0.691 | 0.2628 | No | ||
| 37 | CDC5L | 5339 | 0.658 | 0.2597 | No | ||
| 38 | EIF5 | 5447 | 0.642 | 0.2612 | No | ||
| 39 | POU4F1 | 5907 | 0.573 | 0.2458 | No | ||
| 40 | CHUK | 6349 | 0.506 | 0.2306 | No | ||
| 41 | CCT8 | 6692 | 0.461 | 0.2194 | No | ||
| 42 | ZF | 6874 | 0.435 | 0.2154 | No | ||
| 43 | AGL | 6973 | 0.423 | 0.2151 | No | ||
| 44 | RPL17 | 7860 | 0.306 | 0.1776 | No | ||
| 45 | BZW1 | 8056 | 0.280 | 0.1714 | No | ||
| 46 | RPS6 | 8236 | 0.256 | 0.1657 | No | ||
| 47 | THAP1 | 8360 | 0.238 | 0.1624 | No | ||
| 48 | ZZZ3 | 8870 | 0.166 | 0.1407 | No | ||
| 49 | RAB33B | 9002 | 0.148 | 0.1362 | No | ||
| 50 | LRSAM1 | 10524 | -0.082 | 0.0674 | No | ||
| 51 | ATP5F1 | 10909 | -0.143 | 0.0512 | No | ||
| 52 | ADNP | 10963 | -0.150 | 0.0503 | No | ||
| 53 | DBR1 | 11015 | -0.158 | 0.0495 | No | ||
| 54 | RPL10A | 11098 | -0.168 | 0.0474 | No | ||
| 55 | CHURC1 | 11855 | -0.257 | 0.0153 | No | ||
| 56 | NFYC | 11857 | -0.258 | 0.0178 | No | ||
| 57 | SSR4 | 12064 | -0.279 | 0.0111 | No | ||
| 58 | DCTN2 | 12088 | -0.282 | 0.0128 | No | ||
| 59 | NUDT2 | 12524 | -0.336 | -0.0038 | No | ||
| 60 | MRP63 | 13006 | -0.391 | -0.0220 | No | ||
| 61 | BANP | 13103 | -0.403 | -0.0224 | No | ||
| 62 | CCNJ | 13232 | -0.416 | -0.0242 | No | ||
| 63 | KAZALD1 | 13241 | -0.417 | -0.0204 | No | ||
| 64 | ANAPC4 | 13715 | -0.476 | -0.0374 | No | ||
| 65 | PRDX1 | 13722 | -0.476 | -0.0330 | No | ||
| 66 | MDH2 | 13898 | -0.498 | -0.0361 | No | ||
| 67 | IDH3G | 13949 | -0.503 | -0.0334 | No | ||
| 68 | FSIP1 | 13976 | -0.506 | -0.0297 | No | ||
| 69 | RPL26 | 14312 | -0.546 | -0.0396 | No | ||
| 70 | SLC25A11 | 14463 | -0.564 | -0.0409 | No | ||
| 71 | RPS19 | 14743 | -0.600 | -0.0478 | No | ||
| 72 | DGUOK | 15660 | -0.710 | -0.0828 | No | ||
| 73 | ICMT | 15759 | -0.723 | -0.0801 | No | ||
| 74 | ANKFY1 | 16094 | -0.770 | -0.0879 | No | ||
| 75 | PSMD5 | 16120 | -0.773 | -0.0814 | No | ||
| 76 | VDAC3 | 16246 | -0.789 | -0.0794 | No | ||
| 77 | POLDIP3 | 16352 | -0.804 | -0.0763 | No | ||
| 78 | SUV39H1 | 16413 | -0.811 | -0.0710 | No | ||
| 79 | SUPT5H | 16490 | -0.822 | -0.0664 | No | ||
| 80 | LAMP1 | 16617 | -0.837 | -0.0640 | No | ||
| 81 | RPL12 | 17108 | -0.909 | -0.0774 | No | ||
| 82 | RPS7 | 17325 | -0.941 | -0.0781 | No | ||
| 83 | PRDX4 | 17483 | -0.969 | -0.0757 | No | ||
| 84 | NDUFA11 | 17620 | -0.993 | -0.0722 | No | ||
| 85 | RRAGA | 17730 | -1.012 | -0.0672 | No | ||
| 86 | ZNF346 | 18427 | -1.141 | -0.0879 | No | ||
| 87 | MBTPS1 | 18543 | -1.161 | -0.0817 | No | ||
| 88 | BCAS3 | 18917 | -1.245 | -0.0866 | No | ||
| 89 | CNOT3 | 18921 | -1.245 | -0.0745 | No | ||
| 90 | GAS8 | 19150 | -1.296 | -0.0722 | No | ||
| 91 | SLC25A4 | 19161 | -1.300 | -0.0598 | No | ||
| 92 | COX11 | 19437 | -1.376 | -0.0589 | No | ||
| 93 | WDR6 | 19906 | -1.530 | -0.0653 | No | ||
| 94 | THAP7 | 19967 | -1.554 | -0.0527 | No | ||
| 95 | ATF7 | 20363 | -1.726 | -0.0539 | No | ||
| 96 | ASXL1 | 20608 | -1.851 | -0.0468 | No | ||
| 97 | PFKFB2 | 20777 | -1.955 | -0.0353 | No | ||
| 98 | RPS15A | 20808 | -1.973 | -0.0173 | No | ||
| 99 | MAPK14 | 21120 | -2.209 | -0.0098 | No | ||
| 100 | SAP18 | 21256 | -2.352 | 0.0072 | No | ||
| 101 | ARF3 | 21349 | -2.483 | 0.0274 | No |