Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set02_ATM_minus_versus_BT_ATM_minus |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_pos |
| GeneSet | TCCCRNNRTGC_UNKNOWN |
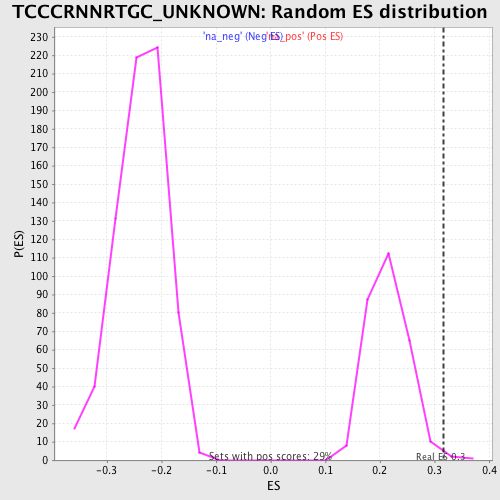
| Enrichment Score (ES) | 0.31670436 |
| Normalized Enrichment Score (NES) | 1.4832892 |
| Nominal p-value | 0.010526316 |
| FDR q-value | 0.20560318 |
| FWER p-Value | 0.815 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | RB1CC1 | 10 | 5.224 | 0.0375 | Yes | ||
| 2 | ORC4L | 29 | 4.329 | 0.0682 | Yes | ||
| 3 | CNOT4 | 51 | 3.789 | 0.0948 | Yes | ||
| 4 | USP15 | 162 | 3.077 | 0.1122 | Yes | ||
| 5 | SNX16 | 174 | 2.997 | 0.1335 | Yes | ||
| 6 | PCM1 | 229 | 2.821 | 0.1515 | Yes | ||
| 7 | CSNK2A1 | 381 | 2.459 | 0.1625 | Yes | ||
| 8 | NEK2 | 382 | 2.458 | 0.1804 | Yes | ||
| 9 | COL4A3BP | 445 | 2.359 | 0.1947 | Yes | ||
| 10 | RBM6 | 696 | 2.060 | 0.1982 | Yes | ||
| 11 | INVS | 807 | 1.960 | 0.2074 | Yes | ||
| 12 | POLK | 872 | 1.906 | 0.2184 | Yes | ||
| 13 | GSPT1 | 975 | 1.835 | 0.2271 | Yes | ||
| 14 | MOSPD2 | 1047 | 1.790 | 0.2368 | Yes | ||
| 15 | ASXL2 | 1084 | 1.767 | 0.2480 | Yes | ||
| 16 | TCTA | 1239 | 1.688 | 0.2533 | Yes | ||
| 17 | CHD4 | 1458 | 1.578 | 0.2547 | Yes | ||
| 18 | CHCHD3 | 1629 | 1.506 | 0.2579 | Yes | ||
| 19 | RAB1A | 1738 | 1.468 | 0.2637 | Yes | ||
| 20 | NSD1 | 1758 | 1.458 | 0.2734 | Yes | ||
| 21 | KIF11 | 1903 | 1.390 | 0.2769 | Yes | ||
| 22 | BSN | 1925 | 1.383 | 0.2860 | Yes | ||
| 23 | VAPA | 2035 | 1.350 | 0.2908 | Yes | ||
| 24 | ZCCHC8 | 2116 | 1.323 | 0.2968 | Yes | ||
| 25 | WTAP | 2281 | 1.267 | 0.2985 | Yes | ||
| 26 | RAD50 | 2391 | 1.227 | 0.3024 | Yes | ||
| 27 | PNRC2 | 2450 | 1.211 | 0.3086 | Yes | ||
| 28 | RHOQ | 2494 | 1.200 | 0.3153 | Yes | ||
| 29 | TIMP4 | 2789 | 1.131 | 0.3101 | Yes | ||
| 30 | SON | 2959 | 1.092 | 0.3103 | Yes | ||
| 31 | NUTF2 | 3125 | 1.054 | 0.3104 | Yes | ||
| 32 | IPO7 | 3399 | 0.991 | 0.3051 | Yes | ||
| 33 | USP16 | 3421 | 0.987 | 0.3113 | Yes | ||
| 34 | ISL1 | 3460 | 0.977 | 0.3167 | Yes | ||
| 35 | TCEB1 | 3612 | 0.946 | 0.3167 | No | ||
| 36 | XPOT | 4230 | 0.834 | 0.2945 | No | ||
| 37 | UBADC1 | 4263 | 0.829 | 0.2990 | No | ||
| 38 | KPNB1 | 4321 | 0.818 | 0.3024 | No | ||
| 39 | GSK3A | 4436 | 0.796 | 0.3029 | No | ||
| 40 | DQX1 | 4481 | 0.789 | 0.3067 | No | ||
| 41 | NAP1L3 | 4493 | 0.788 | 0.3119 | No | ||
| 42 | FUK | 4617 | 0.770 | 0.3119 | No | ||
| 43 | KHSRP | 4642 | 0.766 | 0.3163 | No | ||
| 44 | PLK1 | 5036 | 0.703 | 0.3035 | No | ||
| 45 | EIF5A | 5477 | 0.638 | 0.2879 | No | ||
| 46 | RDH10 | 5499 | 0.634 | 0.2916 | No | ||
| 47 | TRIM23 | 5864 | 0.580 | 0.2791 | No | ||
| 48 | PUM1 | 6066 | 0.550 | 0.2739 | No | ||
| 49 | BRS3 | 6543 | 0.481 | 0.2556 | No | ||
| 50 | PCTK1 | 7312 | 0.378 | 0.2232 | No | ||
| 51 | SNX17 | 7601 | 0.340 | 0.2125 | No | ||
| 52 | COX4I1 | 7960 | 0.293 | 0.1982 | No | ||
| 53 | KLK9 | 8062 | 0.279 | 0.1956 | No | ||
| 54 | UBL3 | 8096 | 0.273 | 0.1961 | No | ||
| 55 | ADSS | 8299 | 0.247 | 0.1886 | No | ||
| 56 | STMN1 | 8643 | 0.198 | 0.1744 | No | ||
| 57 | INSRR | 8818 | 0.173 | 0.1676 | No | ||
| 58 | RPP30 | 8975 | 0.152 | 0.1616 | No | ||
| 59 | SFRS9 | 9439 | 0.083 | 0.1410 | No | ||
| 60 | TTC1 | 9517 | 0.072 | 0.1380 | No | ||
| 61 | IFRD2 | 10127 | -0.023 | 0.1103 | No | ||
| 62 | PRCC | 10704 | -0.108 | 0.0847 | No | ||
| 63 | SNX11 | 10985 | -0.153 | 0.0730 | No | ||
| 64 | IRF2BP1 | 11423 | -0.207 | 0.0544 | No | ||
| 65 | CDK5 | 11504 | -0.217 | 0.0524 | No | ||
| 66 | ACADVL | 12216 | -0.297 | 0.0219 | No | ||
| 67 | MAG | 12268 | -0.303 | 0.0218 | No | ||
| 68 | BUB1B | 12571 | -0.341 | 0.0105 | No | ||
| 69 | VKORC1 | 12594 | -0.343 | 0.0119 | No | ||
| 70 | RPL30 | 12665 | -0.351 | 0.0113 | No | ||
| 71 | ABCB6 | 12927 | -0.383 | 0.0021 | No | ||
| 72 | SFRS1 | 13144 | -0.407 | -0.0048 | No | ||
| 73 | KBTBD7 | 13183 | -0.411 | -0.0036 | No | ||
| 74 | CKAP4 | 13734 | -0.477 | -0.0253 | No | ||
| 75 | RPL7 | 13934 | -0.502 | -0.0307 | No | ||
| 76 | PHACTR3 | 14258 | -0.540 | -0.0416 | No | ||
| 77 | EIF2B4 | 14307 | -0.546 | -0.0398 | No | ||
| 78 | BRMS1 | 14568 | -0.578 | -0.0475 | No | ||
| 79 | SLC4A2 | 14894 | -0.617 | -0.0579 | No | ||
| 80 | GRIN2B | 15010 | -0.629 | -0.0586 | No | ||
| 81 | MRPL24 | 15370 | -0.673 | -0.0702 | No | ||
| 82 | LIG1 | 15585 | -0.699 | -0.0749 | No | ||
| 83 | SMPD3 | 15637 | -0.706 | -0.0721 | No | ||
| 84 | LZTR1 | 15676 | -0.713 | -0.0687 | No | ||
| 85 | TUBGCP3 | 15749 | -0.721 | -0.0667 | No | ||
| 86 | NUDT13 | 15773 | -0.725 | -0.0625 | No | ||
| 87 | FYCO1 | 15852 | -0.735 | -0.0607 | No | ||
| 88 | PPP1R8 | 15890 | -0.741 | -0.0570 | No | ||
| 89 | MIR16 | 16289 | -0.796 | -0.0695 | No | ||
| 90 | PSMD11 | 16972 | -0.887 | -0.0942 | No | ||
| 91 | HSPB9 | 17240 | -0.929 | -0.0997 | No | ||
| 92 | PA2G4 | 17267 | -0.932 | -0.0941 | No | ||
| 93 | NOS3 | 17323 | -0.941 | -0.0898 | No | ||
| 94 | GART | 17331 | -0.942 | -0.0833 | No | ||
| 95 | BET1 | 17400 | -0.954 | -0.0794 | No | ||
| 96 | MBD6 | 17426 | -0.958 | -0.0736 | No | ||
| 97 | RHOA | 17480 | -0.969 | -0.0690 | No | ||
| 98 | PMM2 | 17587 | -0.988 | -0.0667 | No | ||
| 99 | GCN5L2 | 17601 | -0.990 | -0.0601 | No | ||
| 100 | VCL | 18239 | -1.100 | -0.0812 | No | ||
| 101 | PLAGL2 | 18293 | -1.111 | -0.0756 | No | ||
| 102 | MRPL50 | 18459 | -1.145 | -0.0748 | No | ||
| 103 | SCAMP4 | 18863 | -1.232 | -0.0843 | No | ||
| 104 | FBS1 | 19095 | -1.281 | -0.0856 | No | ||
| 105 | EYA4 | 19386 | -1.362 | -0.0890 | No | ||
| 106 | HDAC8 | 19468 | -1.386 | -0.0826 | No | ||
| 107 | TRAF3IP1 | 19632 | -1.434 | -0.0796 | No | ||
| 108 | RASSF6 | 19856 | -1.514 | -0.0788 | No | ||
| 109 | PES1 | 19934 | -1.542 | -0.0711 | No | ||
| 110 | CASC3 | 19977 | -1.557 | -0.0617 | No | ||
| 111 | MORF4L1 | 20258 | -1.680 | -0.0623 | No | ||
| 112 | ASXL1 | 20608 | -1.851 | -0.0649 | No | ||
| 113 | POFUT1 | 21343 | -2.474 | -0.0805 | No | ||
| 114 | POLR1A | 21356 | -2.493 | -0.0629 | No | ||
| 115 | MBP | 21512 | -2.822 | -0.0495 | No | ||
| 116 | NMT1 | 21532 | -2.864 | -0.0295 | No | ||
| 117 | RRS1 | 21621 | -3.150 | -0.0106 | No | ||
| 118 | TLE4 | 21695 | -3.506 | 0.0115 | No |