Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set02_ATM_minus_versus_BT_ATM_minus |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_pos |
| GeneSet | KCCGNSWTTT_UNKNOWN |
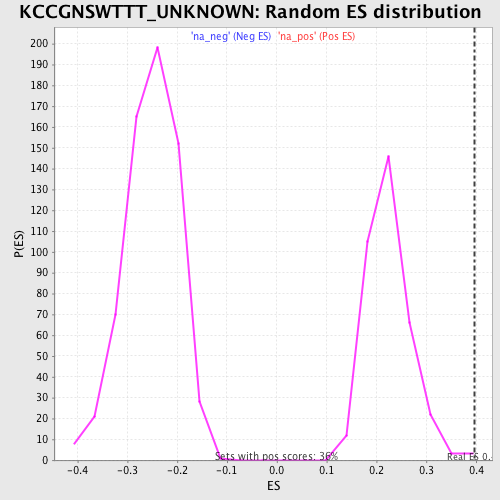
| Enrichment Score (ES) | 0.39729956 |
| Normalized Enrichment Score (NES) | 1.7757716 |
| Nominal p-value | 0.0028011205 |
| FDR q-value | 0.1142375 |
| FWER p-Value | 0.124 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | TAF15 | 72 | 3.634 | 0.0345 | Yes | ||
| 2 | FOXO3A | 134 | 3.219 | 0.0652 | Yes | ||
| 3 | SOS1 | 370 | 2.483 | 0.0803 | Yes | ||
| 4 | PAFAH1B1 | 513 | 2.259 | 0.0973 | Yes | ||
| 5 | OGT | 590 | 2.167 | 0.1164 | Yes | ||
| 6 | ZFP91 | 657 | 2.099 | 0.1352 | Yes | ||
| 7 | SFRS10 | 698 | 2.057 | 0.1548 | Yes | ||
| 8 | USP3 | 729 | 2.028 | 0.1745 | Yes | ||
| 9 | CRK | 753 | 2.008 | 0.1943 | Yes | ||
| 10 | JARID2 | 776 | 1.986 | 0.2140 | Yes | ||
| 11 | PCF11 | 1156 | 1.729 | 0.2147 | Yes | ||
| 12 | PICALM | 1210 | 1.703 | 0.2300 | Yes | ||
| 13 | NCAM1 | 1348 | 1.630 | 0.2407 | Yes | ||
| 14 | LNPEP | 1456 | 1.579 | 0.2522 | Yes | ||
| 15 | MLL5 | 1502 | 1.561 | 0.2664 | Yes | ||
| 16 | YWHAE | 1550 | 1.540 | 0.2803 | Yes | ||
| 17 | LPHN3 | 1559 | 1.538 | 0.2959 | Yes | ||
| 18 | PTBP2 | 1577 | 1.531 | 0.3111 | Yes | ||
| 19 | DMTF1 | 1633 | 1.505 | 0.3242 | Yes | ||
| 20 | CSNK1A1 | 1654 | 1.498 | 0.3389 | Yes | ||
| 21 | HNRPK | 1685 | 1.489 | 0.3530 | Yes | ||
| 22 | RBM4 | 1716 | 1.475 | 0.3670 | Yes | ||
| 23 | SRRM1 | 1904 | 1.390 | 0.3729 | Yes | ||
| 24 | DDX6 | 1948 | 1.376 | 0.3852 | Yes | ||
| 25 | WDR33 | 2155 | 1.312 | 0.3895 | Yes | ||
| 26 | HNRPH1 | 2521 | 1.192 | 0.3852 | Yes | ||
| 27 | POU2F1 | 2833 | 1.121 | 0.3826 | Yes | ||
| 28 | TDRD3 | 3030 | 1.075 | 0.3848 | Yes | ||
| 29 | FBXL11 | 3036 | 1.074 | 0.3958 | Yes | ||
| 30 | SYNCRIP | 3238 | 1.028 | 0.3973 | Yes | ||
| 31 | BCL11A | 3568 | 0.955 | 0.3922 | No | ||
| 32 | CBX3 | 3700 | 0.929 | 0.3959 | No | ||
| 33 | STAG2 | 4119 | 0.852 | 0.3856 | No | ||
| 34 | SFRS3 | 4479 | 0.790 | 0.3774 | No | ||
| 35 | RBPSUH | 4891 | 0.726 | 0.3662 | No | ||
| 36 | EIF4G2 | 5488 | 0.636 | 0.3455 | No | ||
| 37 | CTCF | 5879 | 0.578 | 0.3337 | No | ||
| 38 | CCNA2 | 6116 | 0.542 | 0.3286 | No | ||
| 39 | CUGBP1 | 6374 | 0.503 | 0.3220 | No | ||
| 40 | E2F3 | 7113 | 0.406 | 0.2925 | No | ||
| 41 | HNRPA2B1 | 7468 | 0.357 | 0.2800 | No | ||
| 42 | XPO1 | 7628 | 0.335 | 0.2763 | No | ||
| 43 | PAK7 | 7810 | 0.313 | 0.2712 | No | ||
| 44 | SP1 | 8214 | 0.260 | 0.2555 | No | ||
| 45 | PTMA | 9279 | 0.107 | 0.2080 | No | ||
| 46 | LRRC1 | 9526 | 0.071 | 0.1975 | No | ||
| 47 | JMJD1A | 9795 | 0.028 | 0.1855 | No | ||
| 48 | EIF4A2 | 9987 | -0.002 | 0.1768 | No | ||
| 49 | FOXA2 | 10563 | -0.088 | 0.1514 | No | ||
| 50 | ASCL1 | 10604 | -0.094 | 0.1506 | No | ||
| 51 | ADNP | 10963 | -0.150 | 0.1357 | No | ||
| 52 | DHX15 | 10989 | -0.153 | 0.1362 | No | ||
| 53 | AKAP8 | 11162 | -0.176 | 0.1302 | No | ||
| 54 | NHS | 11723 | -0.241 | 0.1071 | No | ||
| 55 | MAX | 12306 | -0.310 | 0.0837 | No | ||
| 56 | RGL2 | 12606 | -0.345 | 0.0736 | No | ||
| 57 | GNG11 | 13521 | -0.453 | 0.0365 | No | ||
| 58 | CRKL | 13846 | -0.492 | 0.0268 | No | ||
| 59 | PPP2R2B | 14403 | -0.557 | 0.0072 | No | ||
| 60 | IRX5 | 15000 | -0.628 | -0.0135 | No | ||
| 61 | DDX5 | 15849 | -0.734 | -0.0447 | No | ||
| 62 | OTX2 | 16124 | -0.774 | -0.0492 | No | ||
| 63 | CXXC5 | 17224 | -0.927 | -0.0898 | No | ||
| 64 | SPRED2 | 17532 | -0.978 | -0.0936 | No | ||
| 65 | ATP5G2 | 17630 | -0.996 | -0.0877 | No | ||
| 66 | NONO | 17997 | -1.058 | -0.0934 | No | ||
| 67 | DDX17 | 18608 | -1.175 | -0.1091 | No | ||
| 68 | UBC | 18911 | -1.243 | -0.1100 | No | ||
| 69 | CNOT3 | 18921 | -1.245 | -0.0975 | No | ||
| 70 | AP3D1 | 19485 | -1.390 | -0.1087 | No | ||
| 71 | ILF3 | 19614 | -1.429 | -0.0997 | No | ||
| 72 | DAP3 | 19633 | -1.434 | -0.0856 | No | ||
| 73 | AP1G1 | 20050 | -1.590 | -0.0881 | No | ||
| 74 | SF1 | 20461 | -1.773 | -0.0884 | No | ||
| 75 | ENSA | 20479 | -1.781 | -0.0706 | No | ||
| 76 | ACLY | 20502 | -1.795 | -0.0530 | No | ||
| 77 | RFX1 | 20626 | -1.862 | -0.0392 | No | ||
| 78 | SYT11 | 20779 | -1.957 | -0.0258 | No | ||
| 79 | ARMCX2 | 21145 | -2.230 | -0.0193 | No | ||
| 80 | PRICKLE1 | 21424 | -2.616 | -0.0048 | No | ||
| 81 | FAAH | 21484 | -2.754 | 0.0212 | No |