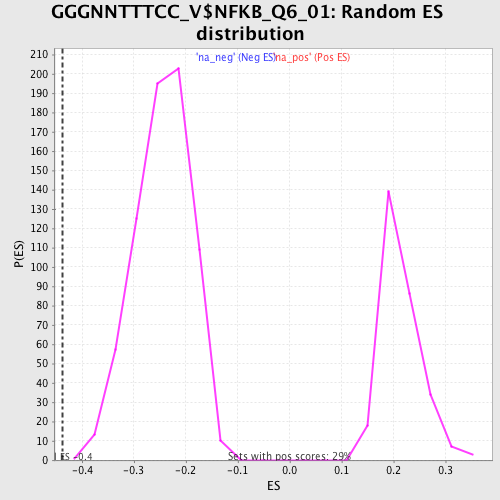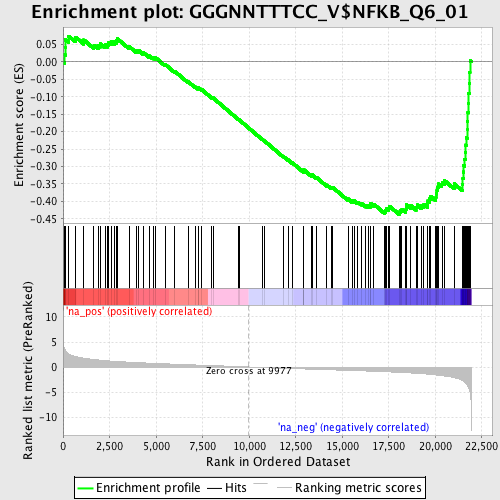
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set02_ATM_minus_versus_BT_ATM_minus |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_neg |
| GeneSet | GGGNNTTTCC_V$NFKB_Q6_01 |
| Enrichment Score (ES) | -0.43748996 |
| Normalized Enrichment Score (NES) | -1.787322 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.014522467 |
| FWER p-Value | 0.152 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | CDC27 | 73 | 3.632 | 0.0215 | No | ||
| 2 | CHD6 | 116 | 3.310 | 0.0422 | No | ||
| 3 | CREB1 | 126 | 3.257 | 0.0641 | No | ||
| 4 | SLC12A2 | 297 | 2.620 | 0.0742 | No | ||
| 5 | RBMS1 | 666 | 2.092 | 0.0717 | No | ||
| 6 | PTGES | 1114 | 1.751 | 0.0632 | No | ||
| 7 | CD86 | 1658 | 1.497 | 0.0486 | No | ||
| 8 | PPP2R5E | 1899 | 1.393 | 0.0471 | No | ||
| 9 | NFAT5 | 1993 | 1.363 | 0.0522 | No | ||
| 10 | BMP2K | 2252 | 1.276 | 0.0491 | No | ||
| 11 | SH3KBP1 | 2392 | 1.226 | 0.0512 | No | ||
| 12 | SOX5 | 2456 | 1.209 | 0.0565 | No | ||
| 13 | BNC2 | 2586 | 1.180 | 0.0587 | No | ||
| 14 | LAMA1 | 2771 | 1.133 | 0.0580 | No | ||
| 15 | CPEB4 | 2861 | 1.114 | 0.0616 | No | ||
| 16 | ALG6 | 2907 | 1.105 | 0.0671 | No | ||
| 17 | RAP2C | 3553 | 0.957 | 0.0441 | No | ||
| 18 | KCNS3 | 3945 | 0.881 | 0.0322 | No | ||
| 19 | SLC6A12 | 4059 | 0.862 | 0.0330 | No | ||
| 20 | NTRK3 | 4304 | 0.821 | 0.0274 | No | ||
| 21 | CCL5 | 4636 | 0.767 | 0.0175 | No | ||
| 22 | IL1RAPL1 | 4833 | 0.734 | 0.0136 | No | ||
| 23 | TUBA3 | 4974 | 0.712 | 0.0120 | No | ||
| 24 | TOM1L1 | 5481 | 0.638 | -0.0068 | No | ||
| 25 | FGF1 | 5996 | 0.558 | -0.0265 | No | ||
| 26 | C1QL1 | 6719 | 0.457 | -0.0564 | No | ||
| 27 | E2F3 | 7113 | 0.406 | -0.0716 | No | ||
| 28 | EDG5 | 7274 | 0.384 | -0.0763 | No | ||
| 29 | HOXA4 | 7275 | 0.384 | -0.0737 | No | ||
| 30 | FUT7 | 7456 | 0.359 | -0.0795 | No | ||
| 31 | RYBP | 7984 | 0.290 | -0.1016 | No | ||
| 32 | LPO | 8053 | 0.280 | -0.1028 | No | ||
| 33 | UBE2M | 9398 | 0.089 | -0.1637 | No | ||
| 34 | HSPG2 | 9466 | 0.079 | -0.1663 | No | ||
| 35 | MSX1 | 10717 | -0.110 | -0.2227 | No | ||
| 36 | MSC | 10845 | -0.131 | -0.2276 | No | ||
| 37 | TRIM47 | 11828 | -0.255 | -0.2708 | No | ||
| 38 | ADMR | 12090 | -0.282 | -0.2809 | No | ||
| 39 | GREM1 | 12314 | -0.311 | -0.2889 | No | ||
| 40 | SLC25A18 | 12913 | -0.381 | -0.3137 | No | ||
| 41 | CNTNAP1 | 12935 | -0.383 | -0.3121 | No | ||
| 42 | BCL6B | 12936 | -0.383 | -0.3094 | No | ||
| 43 | PFN1 | 13354 | -0.432 | -0.3256 | No | ||
| 44 | NRIP2 | 13378 | -0.436 | -0.3236 | No | ||
| 45 | POU2F3 | 13591 | -0.459 | -0.3302 | No | ||
| 46 | UPF2 | 14163 | -0.528 | -0.3527 | No | ||
| 47 | ARID1A | 14401 | -0.557 | -0.3598 | No | ||
| 48 | REL | 14487 | -0.568 | -0.3598 | No | ||
| 49 | TFEC | 15308 | -0.666 | -0.3927 | No | ||
| 50 | GATA4 | 15522 | -0.689 | -0.3978 | No | ||
| 51 | SMPD3 | 15637 | -0.706 | -0.3982 | No | ||
| 52 | FHL3 | 15831 | -0.732 | -0.4020 | No | ||
| 53 | IL6 | 16016 | -0.760 | -0.4052 | No | ||
| 54 | EHD1 | 16254 | -0.790 | -0.4107 | No | ||
| 55 | NFKBIA | 16391 | -0.809 | -0.4113 | No | ||
| 56 | GEM | 16500 | -0.823 | -0.4107 | No | ||
| 57 | UBD | 16505 | -0.824 | -0.4052 | No | ||
| 58 | TNKS1BP1 | 16688 | -0.846 | -0.4078 | No | ||
| 59 | PLXNB1 | 17260 | -0.931 | -0.4275 | No | ||
| 60 | BIRC3 | 17339 | -0.944 | -0.4246 | No | ||
| 61 | WNT10A | 17395 | -0.953 | -0.4206 | No | ||
| 62 | ETV1 | 17507 | -0.973 | -0.4191 | No | ||
| 63 | MAML2 | 17538 | -0.979 | -0.4137 | No | ||
| 64 | IL1RN | 18058 | -1.069 | -0.4302 | Yes | ||
| 65 | RPS6KA4 | 18117 | -1.078 | -0.4255 | Yes | ||
| 66 | IPO4 | 18204 | -1.093 | -0.4219 | Yes | ||
| 67 | UBE2H | 18421 | -1.140 | -0.4240 | Yes | ||
| 68 | TLX1 | 18435 | -1.142 | -0.4168 | Yes | ||
| 69 | FKHL18 | 18450 | -1.144 | -0.4096 | Yes | ||
| 70 | BLR1 | 18680 | -1.189 | -0.4120 | Yes | ||
| 71 | CYLD | 18965 | -1.255 | -0.4164 | Yes | ||
| 72 | CSF1R | 19015 | -1.265 | -0.4100 | Yes | ||
| 73 | DCAMKL1 | 19253 | -1.325 | -0.4118 | Yes | ||
| 74 | TNIP1 | 19385 | -1.362 | -0.4084 | Yes | ||
| 75 | MADCAM1 | 19568 | -1.416 | -0.4071 | Yes | ||
| 76 | TCEA2 | 19600 | -1.424 | -0.3988 | Yes | ||
| 77 | TP53 | 19687 | -1.455 | -0.3927 | Yes | ||
| 78 | MT2A | 19744 | -1.475 | -0.3852 | Yes | ||
| 79 | HIVEP1 | 20031 | -1.583 | -0.3875 | Yes | ||
| 80 | LTB | 20040 | -1.586 | -0.3770 | Yes | ||
| 81 | PCSK2 | 20085 | -1.604 | -0.3680 | Yes | ||
| 82 | TNFSF7 | 20122 | -1.618 | -0.3586 | Yes | ||
| 83 | TPM3 | 20175 | -1.642 | -0.3498 | Yes | ||
| 84 | SLAMF8 | 20358 | -1.724 | -0.3463 | Yes | ||
| 85 | MYST2 | 20483 | -1.782 | -0.3398 | Yes | ||
| 86 | NR2F2 | 21012 | -2.123 | -0.3494 | Yes | ||
| 87 | RASSF2 | 21452 | -2.692 | -0.3511 | Yes | ||
| 88 | SP6 | 21482 | -2.751 | -0.3336 | Yes | ||
| 89 | BATF | 21502 | -2.787 | -0.3154 | Yes | ||
| 90 | STAT6 | 21519 | -2.830 | -0.2968 | Yes | ||
| 91 | JAK3 | 21578 | -3.015 | -0.2788 | Yes | ||
| 92 | NFKB2 | 21615 | -3.137 | -0.2590 | Yes | ||
| 93 | MYADM | 21637 | -3.193 | -0.2381 | Yes | ||
| 94 | IER5 | 21661 | -3.308 | -0.2165 | Yes | ||
| 95 | ENO3 | 21711 | -3.589 | -0.1942 | Yes | ||
| 96 | CD74 | 21718 | -3.626 | -0.1697 | Yes | ||
| 97 | RELB | 21749 | -3.826 | -0.1449 | Yes | ||
| 98 | TNFRSF9 | 21782 | -4.110 | -0.1182 | Yes | ||
| 99 | CCND2 | 21792 | -4.179 | -0.0901 | Yes | ||
| 100 | LASP1 | 21823 | -4.444 | -0.0610 | Yes | ||
| 101 | TNFRSF1B | 21851 | -4.763 | -0.0297 | Yes | ||
| 102 | BCL3 | 21865 | -4.977 | 0.0038 | Yes |