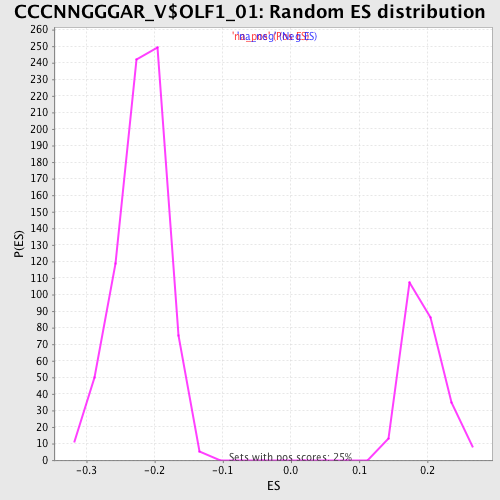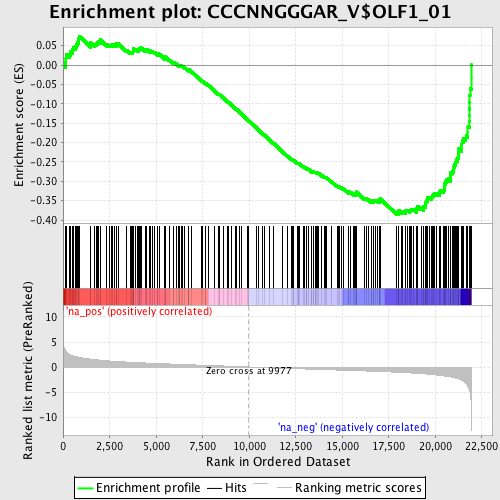
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set02_ATM_minus_versus_BT_ATM_minus |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_neg |
| GeneSet | CCCNNGGGAR_V$OLF1_01 |
| Enrichment Score (ES) | -0.38560495 |
| Normalized Enrichment Score (NES) | -1.7543198 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.01584481 |
| FWER p-Value | 0.231 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | PLAG1 | 128 | 3.249 | 0.0061 | No | ||
| 2 | PPP3CB | 153 | 3.144 | 0.0166 | No | ||
| 3 | IGF2R | 156 | 3.130 | 0.0280 | No | ||
| 4 | SNX13 | 354 | 2.510 | 0.0282 | No | ||
| 5 | MAPKAP1 | 418 | 2.399 | 0.0341 | No | ||
| 6 | ZFHX1B | 495 | 2.285 | 0.0390 | No | ||
| 7 | LPP | 532 | 2.236 | 0.0456 | No | ||
| 8 | LRP1 | 663 | 2.094 | 0.0473 | No | ||
| 9 | BAZ2A | 707 | 2.045 | 0.0529 | No | ||
| 10 | JARID2 | 776 | 1.986 | 0.0571 | No | ||
| 11 | MTSS1 | 825 | 1.936 | 0.0620 | No | ||
| 12 | DPF1 | 846 | 1.922 | 0.0681 | No | ||
| 13 | LRP8 | 859 | 1.912 | 0.0746 | No | ||
| 14 | FGF11 | 1489 | 1.566 | 0.0515 | No | ||
| 15 | PLXDC2 | 1491 | 1.565 | 0.0572 | No | ||
| 16 | REPIN1 | 1711 | 1.477 | 0.0525 | No | ||
| 17 | LUC7L2 | 1778 | 1.447 | 0.0548 | No | ||
| 18 | SLC41A1 | 1832 | 1.422 | 0.0576 | No | ||
| 19 | ARMCX3 | 1884 | 1.401 | 0.0604 | No | ||
| 20 | SDCCAG8 | 1998 | 1.361 | 0.0602 | No | ||
| 21 | TTYH1 | 2003 | 1.360 | 0.0651 | No | ||
| 22 | PCYT1B | 2353 | 1.240 | 0.0536 | No | ||
| 23 | RHOQ | 2494 | 1.200 | 0.0515 | No | ||
| 24 | ATP1A1 | 2584 | 1.180 | 0.0518 | No | ||
| 25 | RORC | 2672 | 1.158 | 0.0521 | No | ||
| 26 | DDR1 | 2758 | 1.137 | 0.0523 | No | ||
| 27 | RAB10 | 2870 | 1.112 | 0.0513 | No | ||
| 28 | UPF3B | 2881 | 1.110 | 0.0549 | No | ||
| 29 | CCDC8 | 2958 | 1.093 | 0.0555 | No | ||
| 30 | GNAO1 | 3428 | 0.985 | 0.0375 | No | ||
| 31 | PRRX1 | 3618 | 0.945 | 0.0323 | No | ||
| 32 | ETV4 | 3689 | 0.931 | 0.0325 | No | ||
| 33 | PTAFR | 3743 | 0.919 | 0.0334 | No | ||
| 34 | ADRB1 | 3755 | 0.917 | 0.0363 | No | ||
| 35 | SPIB | 3763 | 0.916 | 0.0394 | No | ||
| 36 | SP4 | 3782 | 0.911 | 0.0419 | No | ||
| 37 | TIAM1 | 3877 | 0.895 | 0.0409 | No | ||
| 38 | GRIK3 | 4007 | 0.871 | 0.0381 | No | ||
| 39 | STC2 | 4062 | 0.861 | 0.0388 | No | ||
| 40 | HPCA | 4081 | 0.858 | 0.0412 | No | ||
| 41 | STAG2 | 4119 | 0.852 | 0.0426 | No | ||
| 42 | SFRP2 | 4157 | 0.845 | 0.0440 | No | ||
| 43 | SLC25A28 | 4222 | 0.835 | 0.0441 | No | ||
| 44 | HS3ST2 | 4445 | 0.794 | 0.0368 | No | ||
| 45 | H3F3B | 4464 | 0.791 | 0.0389 | No | ||
| 46 | MAGEL2 | 4495 | 0.788 | 0.0405 | No | ||
| 47 | SP3 | 4662 | 0.763 | 0.0356 | No | ||
| 48 | NUP62 | 4702 | 0.756 | 0.0366 | No | ||
| 49 | LHX2 | 4783 | 0.743 | 0.0357 | No | ||
| 50 | TRIM29 | 4887 | 0.727 | 0.0336 | No | ||
| 51 | ECM2 | 5073 | 0.698 | 0.0277 | No | ||
| 52 | PIAS1 | 5080 | 0.698 | 0.0300 | No | ||
| 53 | ELL2 | 5179 | 0.683 | 0.0280 | No | ||
| 54 | DMPK | 5461 | 0.640 | 0.0174 | No | ||
| 55 | MTX1 | 5475 | 0.638 | 0.0191 | No | ||
| 56 | TOM1L1 | 5481 | 0.638 | 0.0213 | No | ||
| 57 | DPYSL5 | 5714 | 0.605 | 0.0128 | No | ||
| 58 | BCL9L | 5910 | 0.572 | 0.0059 | No | ||
| 59 | COL11A2 | 5940 | 0.568 | 0.0067 | No | ||
| 60 | TMEM24 | 6068 | 0.550 | 0.0029 | No | ||
| 61 | DAB1 | 6194 | 0.528 | -0.0009 | No | ||
| 62 | ANXA9 | 6252 | 0.519 | -0.0017 | No | ||
| 63 | GRM7 | 6264 | 0.517 | -0.0003 | No | ||
| 64 | SNCB | 6345 | 0.507 | -0.0021 | No | ||
| 65 | HEBP2 | 6397 | 0.500 | -0.0026 | No | ||
| 66 | IL11 | 6517 | 0.484 | -0.0063 | No | ||
| 67 | PPFIA2 | 6718 | 0.457 | -0.0138 | No | ||
| 68 | C1QL1 | 6719 | 0.457 | -0.0121 | No | ||
| 69 | EVI1 | 6748 | 0.452 | -0.0117 | No | ||
| 70 | ADCYAP1 | 6907 | 0.431 | -0.0174 | No | ||
| 71 | HOXC10 | 7416 | 0.366 | -0.0395 | No | ||
| 72 | CKM | 7500 | 0.353 | -0.0420 | No | ||
| 73 | OPCML | 7654 | 0.332 | -0.0478 | No | ||
| 74 | BAT2 | 7675 | 0.329 | -0.0475 | No | ||
| 75 | EPHB3 | 7818 | 0.312 | -0.0529 | No | ||
| 76 | COL18A1 | 8115 | 0.271 | -0.0655 | No | ||
| 77 | CLDN15 | 8356 | 0.238 | -0.0757 | No | ||
| 78 | GNB2 | 8378 | 0.236 | -0.0758 | No | ||
| 79 | PDGFC | 8386 | 0.235 | -0.0752 | No | ||
| 80 | STIM2 | 8614 | 0.202 | -0.0849 | No | ||
| 81 | FSTL3 | 8826 | 0.172 | -0.0940 | No | ||
| 82 | BMP4 | 8880 | 0.165 | -0.0959 | No | ||
| 83 | MPST | 9021 | 0.144 | -0.1018 | No | ||
| 84 | PTMA | 9279 | 0.107 | -0.1132 | No | ||
| 85 | THBS3 | 9303 | 0.104 | -0.1139 | No | ||
| 86 | FBXO11 | 9480 | 0.077 | -0.1217 | No | ||
| 87 | EPN3 | 9601 | 0.060 | -0.1270 | No | ||
| 88 | DTX3 | 9923 | 0.007 | -0.1417 | No | ||
| 89 | MDGA1 | 9938 | 0.005 | -0.1424 | No | ||
| 90 | RAB39 | 9957 | 0.003 | -0.1432 | No | ||
| 91 | C1QTNF1 | 10377 | -0.061 | -0.1622 | No | ||
| 92 | SLC30A5 | 10473 | -0.075 | -0.1663 | No | ||
| 93 | TST | 10706 | -0.109 | -0.1766 | No | ||
| 94 | AAMP | 10798 | -0.122 | -0.1804 | No | ||
| 95 | SGK2 | 11096 | -0.167 | -0.1934 | No | ||
| 96 | ALOX12B | 11291 | -0.192 | -0.2016 | No | ||
| 97 | SMARCE1 | 11786 | -0.248 | -0.2235 | No | ||
| 98 | PDAP1 | 12067 | -0.280 | -0.2353 | No | ||
| 99 | MAG | 12268 | -0.303 | -0.2434 | No | ||
| 100 | GREM1 | 12314 | -0.311 | -0.2443 | No | ||
| 101 | APBA1 | 12379 | -0.318 | -0.2461 | No | ||
| 102 | CHAT | 12400 | -0.320 | -0.2458 | No | ||
| 103 | SSH2 | 12581 | -0.342 | -0.2529 | No | ||
| 104 | ATF5 | 12624 | -0.348 | -0.2535 | No | ||
| 105 | LIN28 | 12659 | -0.351 | -0.2538 | No | ||
| 106 | FBXL19 | 12676 | -0.352 | -0.2532 | No | ||
| 107 | TENC1 | 12903 | -0.380 | -0.2622 | No | ||
| 108 | TBX21 | 12958 | -0.386 | -0.2633 | No | ||
| 109 | NEUROD2 | 13050 | -0.397 | -0.2660 | No | ||
| 110 | BANP | 13103 | -0.403 | -0.2669 | No | ||
| 111 | PRDM10 | 13184 | -0.411 | -0.2691 | No | ||
| 112 | NR2E1 | 13365 | -0.434 | -0.2758 | No | ||
| 113 | ADAMTS4 | 13367 | -0.435 | -0.2742 | No | ||
| 114 | PDZRN4 | 13432 | -0.443 | -0.2756 | No | ||
| 115 | THRA | 13450 | -0.445 | -0.2747 | No | ||
| 116 | UNG2 | 13534 | -0.455 | -0.2768 | No | ||
| 117 | SYTL2 | 13556 | -0.456 | -0.2761 | No | ||
| 118 | WDR13 | 13620 | -0.463 | -0.2773 | No | ||
| 119 | FKBP2 | 13692 | -0.473 | -0.2788 | No | ||
| 120 | SCG3 | 13741 | -0.478 | -0.2793 | No | ||
| 121 | ADCY4 | 13899 | -0.498 | -0.2847 | No | ||
| 122 | PTHLH | 14038 | -0.513 | -0.2891 | No | ||
| 123 | DUSP22 | 14089 | -0.518 | -0.2895 | No | ||
| 124 | EFS | 14130 | -0.523 | -0.2895 | No | ||
| 125 | ARID1A | 14401 | -0.557 | -0.2998 | No | ||
| 126 | LRRC20 | 14722 | -0.598 | -0.3124 | No | ||
| 127 | GIPC3 | 14804 | -0.607 | -0.3139 | No | ||
| 128 | LRFN3 | 14841 | -0.611 | -0.3133 | No | ||
| 129 | GRIN2D | 14936 | -0.622 | -0.3153 | No | ||
| 130 | CALM1 | 15082 | -0.637 | -0.3196 | No | ||
| 131 | UBE2N | 15327 | -0.668 | -0.3284 | No | ||
| 132 | NDUFS2 | 15358 | -0.672 | -0.3273 | No | ||
| 133 | FLT1 | 15434 | -0.681 | -0.3282 | No | ||
| 134 | GPM6B | 15593 | -0.700 | -0.3329 | No | ||
| 135 | SMPD3 | 15637 | -0.706 | -0.3323 | No | ||
| 136 | BAI2 | 15719 | -0.717 | -0.3334 | No | ||
| 137 | GRIK1 | 15748 | -0.721 | -0.3320 | No | ||
| 138 | DLL4 | 15750 | -0.721 | -0.3294 | No | ||
| 139 | ATP2B3 | 15767 | -0.724 | -0.3275 | No | ||
| 140 | AKAP12 | 16182 | -0.782 | -0.3437 | No | ||
| 141 | CNTN2 | 16277 | -0.794 | -0.3451 | No | ||
| 142 | NFKBIA | 16391 | -0.809 | -0.3473 | No | ||
| 143 | CD79B | 16562 | -0.830 | -0.3521 | No | ||
| 144 | CHL1 | 16583 | -0.833 | -0.3499 | No | ||
| 145 | KCND1 | 16675 | -0.845 | -0.3510 | No | ||
| 146 | TNKS1BP1 | 16688 | -0.846 | -0.3484 | No | ||
| 147 | NEO1 | 16777 | -0.858 | -0.3493 | No | ||
| 148 | CBX6 | 16868 | -0.871 | -0.3502 | No | ||
| 149 | ZDHHC14 | 16987 | -0.889 | -0.3524 | No | ||
| 150 | CHCHD7 | 16994 | -0.890 | -0.3494 | No | ||
| 151 | WNT7B | 16996 | -0.890 | -0.3462 | No | ||
| 152 | HOXA11 | 17053 | -0.901 | -0.3454 | No | ||
| 153 | ABI3 | 17927 | -1.047 | -0.3817 | Yes | ||
| 154 | ENPP2 | 18000 | -1.058 | -0.3812 | Yes | ||
| 155 | ELAVL3 | 18006 | -1.060 | -0.3775 | Yes | ||
| 156 | MYOZ1 | 18042 | -1.066 | -0.3752 | Yes | ||
| 157 | HIC2 | 18205 | -1.093 | -0.3786 | Yes | ||
| 158 | FCHSD2 | 18256 | -1.104 | -0.3768 | Yes | ||
| 159 | GNGT2 | 18408 | -1.137 | -0.3796 | Yes | ||
| 160 | LRRTM3 | 18416 | -1.139 | -0.3757 | Yes | ||
| 161 | GCNT2 | 18491 | -1.153 | -0.3749 | Yes | ||
| 162 | POGK | 18592 | -1.172 | -0.3752 | Yes | ||
| 163 | MESDC1 | 18678 | -1.189 | -0.3747 | Yes | ||
| 164 | CDON | 18713 | -1.197 | -0.3719 | Yes | ||
| 165 | JMJD1B | 18811 | -1.220 | -0.3718 | Yes | ||
| 166 | GIT1 | 18987 | -1.260 | -0.3753 | Yes | ||
| 167 | DLX1 | 18995 | -1.261 | -0.3709 | Yes | ||
| 168 | OPA3 | 19031 | -1.268 | -0.3679 | Yes | ||
| 169 | CHD5 | 19049 | -1.271 | -0.3640 | Yes | ||
| 170 | RHOG | 19258 | -1.326 | -0.3687 | Yes | ||
| 171 | HR | 19382 | -1.361 | -0.3693 | Yes | ||
| 172 | TNIP1 | 19385 | -1.362 | -0.3644 | Yes | ||
| 173 | UBAP1 | 19462 | -1.384 | -0.3628 | Yes | ||
| 174 | ABR | 19483 | -1.389 | -0.3586 | Yes | ||
| 175 | CA2 | 19489 | -1.391 | -0.3537 | Yes | ||
| 176 | MAGED2 | 19536 | -1.407 | -0.3506 | Yes | ||
| 177 | MYOHD1 | 19560 | -1.414 | -0.3465 | Yes | ||
| 178 | MADCAM1 | 19568 | -1.416 | -0.3416 | Yes | ||
| 179 | RNF44 | 19686 | -1.455 | -0.3416 | Yes | ||
| 180 | ICA1 | 19782 | -1.491 | -0.3405 | Yes | ||
| 181 | ROBO3 | 19824 | -1.505 | -0.3368 | Yes | ||
| 182 | NCDN | 19901 | -1.528 | -0.3347 | Yes | ||
| 183 | VAT1 | 19947 | -1.547 | -0.3311 | Yes | ||
| 184 | RBM10 | 20057 | -1.593 | -0.3302 | Yes | ||
| 185 | NF2 | 20221 | -1.662 | -0.3316 | Yes | ||
| 186 | TNNT3 | 20236 | -1.669 | -0.3261 | Yes | ||
| 187 | HCST | 20297 | -1.698 | -0.3226 | Yes | ||
| 188 | TRIB2 | 20441 | -1.762 | -0.3227 | Yes | ||
| 189 | PSME3 | 20480 | -1.781 | -0.3179 | Yes | ||
| 190 | TPM1 | 20488 | -1.784 | -0.3117 | Yes | ||
| 191 | H1F0 | 20504 | -1.798 | -0.3057 | Yes | ||
| 192 | PSCD3 | 20549 | -1.819 | -0.3010 | Yes | ||
| 193 | LIMK2 | 20599 | -1.846 | -0.2965 | Yes | ||
| 194 | CHRDL1 | 20693 | -1.905 | -0.2938 | Yes | ||
| 195 | FOXJ2 | 20816 | -1.975 | -0.2921 | Yes | ||
| 196 | BAHD1 | 20822 | -1.978 | -0.2850 | Yes | ||
| 197 | TRIM8 | 20836 | -1.988 | -0.2783 | Yes | ||
| 198 | CYB561D1 | 20899 | -2.031 | -0.2737 | Yes | ||
| 199 | SLC25A14 | 20971 | -2.084 | -0.2693 | Yes | ||
| 200 | TRAF4 | 21000 | -2.116 | -0.2628 | Yes | ||
| 201 | ELK3 | 21026 | -2.132 | -0.2561 | Yes | ||
| 202 | AXL | 21095 | -2.192 | -0.2511 | Yes | ||
| 203 | ICAM1 | 21114 | -2.204 | -0.2438 | Yes | ||
| 204 | UBTF | 21212 | -2.303 | -0.2398 | Yes | ||
| 205 | ETV6 | 21235 | -2.325 | -0.2323 | Yes | ||
| 206 | GRK5 | 21250 | -2.344 | -0.2243 | Yes | ||
| 207 | JMJD2A | 21267 | -2.363 | -0.2163 | Yes | ||
| 208 | CD72 | 21419 | -2.609 | -0.2137 | Yes | ||
| 209 | COL23A1 | 21422 | -2.614 | -0.2041 | Yes | ||
| 210 | SLC9A6 | 21433 | -2.647 | -0.1948 | Yes | ||
| 211 | ZBTB9 | 21520 | -2.835 | -0.1883 | Yes | ||
| 212 | VASP | 21648 | -3.238 | -0.1823 | Yes | ||
| 213 | RGS3 | 21752 | -3.840 | -0.1729 | Yes | ||
| 214 | DUSP4 | 21754 | -3.844 | -0.1588 | Yes | ||
| 215 | BCOR | 21817 | -4.413 | -0.1454 | Yes | ||
| 216 | LASP1 | 21823 | -4.444 | -0.1292 | Yes | ||
| 217 | NDFIP1 | 21826 | -4.492 | -0.1128 | Yes | ||
| 218 | IL4I1 | 21849 | -4.754 | -0.0963 | Yes | ||
| 219 | PCBP4 | 21856 | -4.849 | -0.0787 | Yes | ||
| 220 | POU2AF1 | 21866 | -4.982 | -0.0607 | Yes | ||
| 221 | PACSIN1 | 21941 | -8.633 | -0.0324 | Yes | ||
| 222 | EGR1 | 21942 | -8.847 | 0.0002 | Yes |