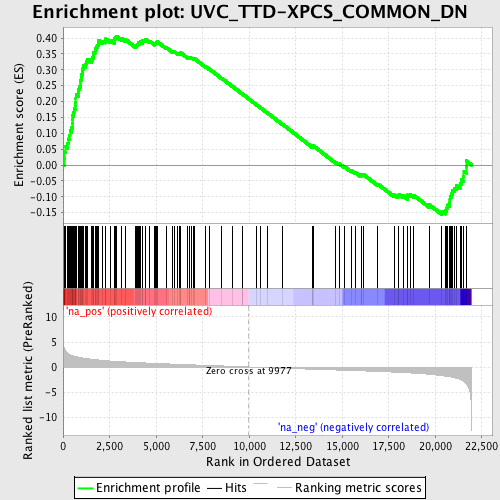
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set02_ATM_minus_versus_BT_ATM_minus |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_pos |
| GeneSet | UVC_TTD-XPCS_COMMON_DN |
| Enrichment Score (ES) | 0.40610355 |
| Normalized Enrichment Score (NES) | 1.9252476 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.029557955 |
| FWER p-Value | 0.498 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | CNOT4 | 51 | 3.789 | 0.0200 | Yes | ||
| 2 | ZBTB20 | 58 | 3.751 | 0.0418 | Yes | ||
| 3 | PIP5K3 | 108 | 3.370 | 0.0593 | Yes | ||
| 4 | FUT8 | 246 | 2.749 | 0.0692 | Yes | ||
| 5 | THRAP1 | 301 | 2.610 | 0.0821 | Yes | ||
| 6 | ATRX | 361 | 2.498 | 0.0941 | Yes | ||
| 7 | SOS1 | 370 | 2.483 | 0.1083 | Yes | ||
| 8 | NCOA1 | 470 | 2.323 | 0.1175 | Yes | ||
| 9 | ZFHX1B | 495 | 2.285 | 0.1298 | Yes | ||
| 10 | KLF6 | 502 | 2.271 | 0.1429 | Yes | ||
| 11 | RUNX1 | 523 | 2.244 | 0.1552 | Yes | ||
| 12 | ZNF292 | 556 | 2.198 | 0.1667 | Yes | ||
| 13 | OGT | 590 | 2.167 | 0.1779 | Yes | ||
| 14 | FALZ | 671 | 2.086 | 0.1865 | Yes | ||
| 15 | BAZ2B | 677 | 2.083 | 0.1985 | Yes | ||
| 16 | UBR2 | 688 | 2.069 | 0.2102 | Yes | ||
| 17 | MARCH7 | 692 | 2.062 | 0.2222 | Yes | ||
| 18 | EDD1 | 813 | 1.953 | 0.2282 | Yes | ||
| 19 | ASCC3 | 844 | 1.922 | 0.2382 | Yes | ||
| 20 | CASK | 873 | 1.906 | 0.2481 | Yes | ||
| 21 | CENTB2 | 933 | 1.865 | 0.2564 | Yes | ||
| 22 | CENPA | 937 | 1.859 | 0.2672 | Yes | ||
| 23 | GAPVD1 | 966 | 1.842 | 0.2767 | Yes | ||
| 24 | GOLGA4 | 1001 | 1.819 | 0.2859 | Yes | ||
| 25 | MTF2 | 1053 | 1.786 | 0.2940 | Yes | ||
| 26 | ARHGAP12 | 1064 | 1.777 | 0.3040 | Yes | ||
| 27 | NUP98 | 1105 | 1.755 | 0.3125 | Yes | ||
| 28 | MBNL1 | 1213 | 1.702 | 0.3176 | Yes | ||
| 29 | APPBP2 | 1235 | 1.689 | 0.3266 | Yes | ||
| 30 | HIRA | 1291 | 1.658 | 0.3339 | Yes | ||
| 31 | RBM39 | 1521 | 1.554 | 0.3325 | Yes | ||
| 32 | KLF7 | 1567 | 1.535 | 0.3395 | Yes | ||
| 33 | ZNF638 | 1625 | 1.508 | 0.3457 | Yes | ||
| 34 | PRKACB | 1648 | 1.500 | 0.3536 | Yes | ||
| 35 | ORC2L | 1715 | 1.475 | 0.3592 | Yes | ||
| 36 | ZMYND11 | 1735 | 1.469 | 0.3670 | Yes | ||
| 37 | ATXN1 | 1811 | 1.433 | 0.3720 | Yes | ||
| 38 | PPAP2B | 1860 | 1.410 | 0.3781 | Yes | ||
| 39 | PPP2R5E | 1899 | 1.393 | 0.3845 | Yes | ||
| 40 | BLM | 1909 | 1.389 | 0.3923 | Yes | ||
| 41 | PDLIM5 | 2142 | 1.316 | 0.3894 | Yes | ||
| 42 | BMP2K | 2252 | 1.276 | 0.3919 | Yes | ||
| 43 | SLC25A13 | 2299 | 1.260 | 0.3972 | Yes | ||
| 44 | PCTK2 | 2547 | 1.187 | 0.3929 | Yes | ||
| 45 | CLASP1 | 2741 | 1.140 | 0.3907 | Yes | ||
| 46 | CDC2L5 | 2744 | 1.140 | 0.3974 | Yes | ||
| 47 | LASS6 | 2792 | 1.130 | 0.4019 | Yes | ||
| 48 | LARP5 | 2844 | 1.119 | 0.4061 | Yes | ||
| 49 | MAP4K5 | 3156 | 1.048 | 0.3980 | No | ||
| 50 | NFIB | 3359 | 0.999 | 0.3946 | No | ||
| 51 | TIAM1 | 3877 | 0.895 | 0.3762 | No | ||
| 52 | NUP153 | 3954 | 0.880 | 0.3779 | No | ||
| 53 | PTPN2 | 4011 | 0.871 | 0.3805 | No | ||
| 54 | MALT1 | 4026 | 0.868 | 0.3849 | No | ||
| 55 | UGCG | 4120 | 0.851 | 0.3857 | No | ||
| 56 | TIPARP | 4146 | 0.848 | 0.3895 | No | ||
| 57 | CRADD | 4268 | 0.828 | 0.3888 | No | ||
| 58 | PHLPPL | 4272 | 0.827 | 0.3936 | No | ||
| 59 | ATP13A3 | 4402 | 0.803 | 0.3924 | No | ||
| 60 | MTAP | 4427 | 0.798 | 0.3960 | No | ||
| 61 | GNAI1 | 4650 | 0.765 | 0.3903 | No | ||
| 62 | PKP4 | 4921 | 0.721 | 0.3822 | No | ||
| 63 | SMCHD1 | 4976 | 0.711 | 0.3839 | No | ||
| 64 | RASA1 | 5023 | 0.704 | 0.3859 | No | ||
| 65 | ROR1 | 5062 | 0.700 | 0.3883 | No | ||
| 66 | UST | 5553 | 0.627 | 0.3695 | No | ||
| 67 | KNTC2 | 5865 | 0.580 | 0.3587 | No | ||
| 68 | KIF2A | 5971 | 0.564 | 0.3572 | No | ||
| 69 | TMEM131 | 6171 | 0.532 | 0.3512 | No | ||
| 70 | RGS20 | 6246 | 0.520 | 0.3509 | No | ||
| 71 | ARIH1 | 6312 | 0.511 | 0.3509 | No | ||
| 72 | ID1 | 6328 | 0.509 | 0.3532 | No | ||
| 73 | FNBP1L | 6685 | 0.461 | 0.3396 | No | ||
| 74 | HOXB2 | 6796 | 0.445 | 0.3372 | No | ||
| 75 | PPP2R3A | 6816 | 0.443 | 0.3389 | No | ||
| 76 | GULP1 | 6910 | 0.430 | 0.3372 | No | ||
| 77 | ACSL3 | 7024 | 0.417 | 0.3345 | No | ||
| 78 | EDG2 | 7070 | 0.410 | 0.3348 | No | ||
| 79 | ANKRD28 | 7662 | 0.330 | 0.3097 | No | ||
| 80 | CXCL12 | 7857 | 0.306 | 0.3026 | No | ||
| 81 | CDK8 | 8515 | 0.215 | 0.2738 | No | ||
| 82 | BAZ1A | 9115 | 0.131 | 0.2471 | No | ||
| 83 | MNAT1 | 9623 | 0.055 | 0.2242 | No | ||
| 84 | SOS2 | 9624 | 0.055 | 0.2245 | No | ||
| 85 | KLF10 | 10404 | -0.066 | 0.1892 | No | ||
| 86 | KIF14 | 10611 | -0.095 | 0.1803 | No | ||
| 87 | DHX15 | 10989 | -0.153 | 0.1639 | No | ||
| 88 | NAV3 | 11788 | -0.248 | 0.1288 | No | ||
| 89 | MELK | 13403 | -0.439 | 0.0574 | No | ||
| 90 | SHB | 13406 | -0.439 | 0.0599 | No | ||
| 91 | GBE1 | 13438 | -0.444 | 0.0611 | No | ||
| 92 | MAP2K4 | 14640 | -0.588 | 0.0096 | No | ||
| 93 | TRIM2 | 14837 | -0.610 | 0.0042 | No | ||
| 94 | GLRB | 15131 | -0.643 | -0.0055 | No | ||
| 95 | DPYD | 15490 | -0.686 | -0.0179 | No | ||
| 96 | FARS2 | 15704 | -0.716 | -0.0234 | No | ||
| 97 | NRG1 | 16010 | -0.759 | -0.0329 | No | ||
| 98 | INPP5F | 16043 | -0.763 | -0.0299 | No | ||
| 99 | MARK3 | 16118 | -0.773 | -0.0287 | No | ||
| 100 | ITSN1 | 16914 | -0.879 | -0.0600 | No | ||
| 101 | SERPINE1 | 17791 | -1.020 | -0.0941 | No | ||
| 102 | SKIV2L2 | 17994 | -1.057 | -0.0972 | No | ||
| 103 | DOCK4 | 18046 | -1.067 | -0.0932 | No | ||
| 104 | DCUN1D4 | 18264 | -1.105 | -0.0967 | No | ||
| 105 | CREB5 | 18507 | -1.155 | -0.1010 | No | ||
| 106 | E2F5 | 18509 | -1.155 | -0.0942 | No | ||
| 107 | TSC22D2 | 18644 | -1.183 | -0.0934 | No | ||
| 108 | GTF2F2 | 18850 | -1.229 | -0.0955 | No | ||
| 109 | AUH | 19679 | -1.451 | -0.1250 | No | ||
| 110 | HOMER1 | 20351 | -1.719 | -0.1456 | No | ||
| 111 | SEPT9 | 20541 | -1.817 | -0.1436 | No | ||
| 112 | ASXL1 | 20608 | -1.851 | -0.1357 | No | ||
| 113 | SLC25A12 | 20633 | -1.866 | -0.1258 | No | ||
| 114 | GRLF1 | 20741 | -1.931 | -0.1194 | No | ||
| 115 | MYO1B | 20786 | -1.959 | -0.1098 | No | ||
| 116 | MTMR3 | 20790 | -1.964 | -0.0984 | No | ||
| 117 | SWAP70 | 20889 | -2.026 | -0.0910 | No | ||
| 118 | DOPEY2 | 20921 | -2.050 | -0.0804 | No | ||
| 119 | ACVR1 | 21020 | -2.130 | -0.0723 | No | ||
| 120 | TRAK1 | 21118 | -2.208 | -0.0638 | No | ||
| 121 | BHLHB2 | 21329 | -2.459 | -0.0589 | No | ||
| 122 | IGF1R | 21392 | -2.559 | -0.0467 | No | ||
| 123 | SMAD3 | 21501 | -2.785 | -0.0353 | No | ||
| 124 | TRAM2 | 21539 | -2.875 | -0.0200 | No | ||
| 125 | ZHX2 | 21654 | -3.267 | -0.0060 | No | ||
| 126 | MGLL | 21658 | -3.302 | 0.0132 | No |