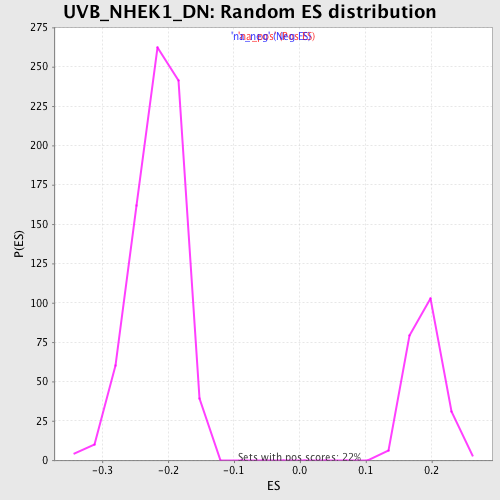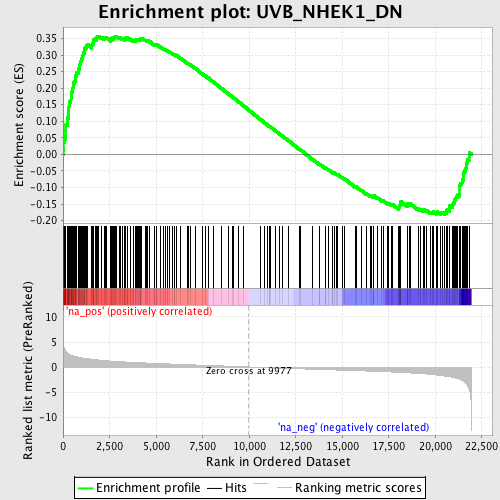
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set02_ATM_minus_versus_BT_ATM_minus |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_pos |
| GeneSet | UVB_NHEK1_DN |
| Enrichment Score (ES) | 0.35712892 |
| Normalized Enrichment Score (NES) | 1.8779092 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.040953558 |
| FWER p-Value | 0.665 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | NIPBL | 14 | 4.794 | 0.0134 | Yes | ||
| 2 | SMC5 | 31 | 4.252 | 0.0251 | Yes | ||
| 3 | SMC4 | 34 | 4.099 | 0.0370 | Yes | ||
| 4 | CNOT4 | 51 | 3.789 | 0.0474 | Yes | ||
| 5 | PIP5K3 | 108 | 3.370 | 0.0547 | Yes | ||
| 6 | FOXO3A | 134 | 3.219 | 0.0630 | Yes | ||
| 7 | NPAT | 138 | 3.201 | 0.0722 | Yes | ||
| 8 | TNFAIP8 | 149 | 3.164 | 0.0810 | Yes | ||
| 9 | MTM1 | 152 | 3.155 | 0.0901 | Yes | ||
| 10 | RABGAP1L | 230 | 2.821 | 0.0949 | Yes | ||
| 11 | POLD3 | 240 | 2.785 | 0.1026 | Yes | ||
| 12 | BRD8 | 243 | 2.758 | 0.1106 | Yes | ||
| 13 | AFF1 | 274 | 2.679 | 0.1171 | Yes | ||
| 14 | SPOP | 285 | 2.654 | 0.1244 | Yes | ||
| 15 | IRAK1BP1 | 296 | 2.621 | 0.1316 | Yes | ||
| 16 | NRIP1 | 304 | 2.608 | 0.1389 | Yes | ||
| 17 | ATP2B1 | 315 | 2.577 | 0.1460 | Yes | ||
| 18 | ZNF148 | 321 | 2.566 | 0.1533 | Yes | ||
| 19 | ATRX | 361 | 2.498 | 0.1588 | Yes | ||
| 20 | DOPEY1 | 415 | 2.402 | 0.1634 | Yes | ||
| 21 | REV3L | 423 | 2.391 | 0.1701 | Yes | ||
| 22 | ATF2 | 444 | 2.361 | 0.1761 | Yes | ||
| 23 | AKAP9 | 469 | 2.324 | 0.1818 | Yes | ||
| 24 | PUM2 | 472 | 2.322 | 0.1885 | Yes | ||
| 25 | PAFAH1B1 | 513 | 2.259 | 0.1932 | Yes | ||
| 26 | CREBBP | 519 | 2.250 | 0.1996 | Yes | ||
| 27 | LPP | 532 | 2.236 | 0.2056 | Yes | ||
| 28 | ELF1 | 551 | 2.205 | 0.2112 | Yes | ||
| 29 | FNDC3A | 557 | 2.196 | 0.2174 | Yes | ||
| 30 | BAZ1B | 613 | 2.144 | 0.2212 | Yes | ||
| 31 | SACS | 648 | 2.112 | 0.2258 | Yes | ||
| 32 | FALZ | 671 | 2.086 | 0.2309 | Yes | ||
| 33 | BAZ2B | 677 | 2.083 | 0.2368 | Yes | ||
| 34 | SSBP2 | 701 | 2.051 | 0.2417 | Yes | ||
| 35 | KIF23 | 717 | 2.037 | 0.2470 | Yes | ||
| 36 | USP1 | 805 | 1.961 | 0.2487 | Yes | ||
| 37 | EDD1 | 813 | 1.953 | 0.2541 | Yes | ||
| 38 | TLK1 | 843 | 1.923 | 0.2584 | Yes | ||
| 39 | CENPF | 863 | 1.909 | 0.2631 | Yes | ||
| 40 | AHR | 871 | 1.907 | 0.2684 | Yes | ||
| 41 | JARID1A | 904 | 1.880 | 0.2724 | Yes | ||
| 42 | CENPA | 937 | 1.859 | 0.2764 | Yes | ||
| 43 | TAF11 | 957 | 1.849 | 0.2809 | Yes | ||
| 44 | GAPVD1 | 966 | 1.842 | 0.2860 | Yes | ||
| 45 | PHF3 | 1006 | 1.813 | 0.2895 | Yes | ||
| 46 | HELZ | 1042 | 1.793 | 0.2931 | Yes | ||
| 47 | WAPAL | 1057 | 1.782 | 0.2977 | Yes | ||
| 48 | WDR37 | 1071 | 1.774 | 0.3023 | Yes | ||
| 49 | RNF13 | 1076 | 1.773 | 0.3073 | Yes | ||
| 50 | TOP2A | 1137 | 1.740 | 0.3096 | Yes | ||
| 51 | PTPN3 | 1144 | 1.736 | 0.3144 | Yes | ||
| 52 | PCF11 | 1156 | 1.729 | 0.3190 | Yes | ||
| 53 | PLK4 | 1193 | 1.711 | 0.3224 | Yes | ||
| 54 | APPBP2 | 1235 | 1.689 | 0.3254 | Yes | ||
| 55 | EFNB2 | 1280 | 1.663 | 0.3283 | Yes | ||
| 56 | VPS13A | 1327 | 1.641 | 0.3310 | Yes | ||
| 57 | KLF9 | 1543 | 1.544 | 0.3256 | Yes | ||
| 58 | USP6NL | 1547 | 1.542 | 0.3300 | Yes | ||
| 59 | KLF7 | 1567 | 1.535 | 0.3336 | Yes | ||
| 60 | SLC25A36 | 1609 | 1.516 | 0.3361 | Yes | ||
| 61 | ZNF638 | 1625 | 1.508 | 0.3399 | Yes | ||
| 62 | DMTF1 | 1633 | 1.505 | 0.3439 | Yes | ||
| 63 | BACH1 | 1650 | 1.500 | 0.3476 | Yes | ||
| 64 | ORC2L | 1715 | 1.475 | 0.3490 | Yes | ||
| 65 | ZCCHC11 | 1803 | 1.437 | 0.3492 | Yes | ||
| 66 | CENPC1 | 1816 | 1.429 | 0.3528 | Yes | ||
| 67 | CNOT2 | 1825 | 1.425 | 0.3566 | Yes | ||
| 68 | KIF11 | 1903 | 1.390 | 0.3571 | Yes | ||
| 69 | WDFY3 | 2082 | 1.334 | 0.3528 | No | ||
| 70 | HMGA2 | 2236 | 1.280 | 0.3495 | No | ||
| 71 | TMCC1 | 2259 | 1.275 | 0.3523 | No | ||
| 72 | SLBP | 2323 | 1.250 | 0.3530 | No | ||
| 73 | RSBN1 | 2565 | 1.184 | 0.3454 | No | ||
| 74 | CEP350 | 2589 | 1.179 | 0.3478 | No | ||
| 75 | MYST3 | 2616 | 1.171 | 0.3500 | No | ||
| 76 | CEP170 | 2651 | 1.164 | 0.3519 | No | ||
| 77 | AUTS2 | 2733 | 1.143 | 0.3515 | No | ||
| 78 | CLASP1 | 2741 | 1.140 | 0.3545 | No | ||
| 79 | LASS6 | 2792 | 1.130 | 0.3555 | No | ||
| 80 | CHD1 | 2850 | 1.117 | 0.3562 | No | ||
| 81 | DYRK2 | 3002 | 1.082 | 0.3524 | No | ||
| 82 | MTIF2 | 3065 | 1.068 | 0.3526 | No | ||
| 83 | TLK2 | 3164 | 1.046 | 0.3512 | No | ||
| 84 | RAPGEF5 | 3312 | 1.010 | 0.3474 | No | ||
| 85 | ABL1 | 3313 | 1.010 | 0.3503 | No | ||
| 86 | PPP1R12A | 3345 | 1.003 | 0.3519 | No | ||
| 87 | DIDO1 | 3435 | 0.984 | 0.3506 | No | ||
| 88 | CEBPD | 3462 | 0.977 | 0.3523 | No | ||
| 89 | SFRS2IP | 3632 | 0.942 | 0.3473 | No | ||
| 90 | ARID3A | 3794 | 0.910 | 0.3425 | No | ||
| 91 | PHC2 | 3801 | 0.909 | 0.3449 | No | ||
| 92 | CPEB3 | 3883 | 0.894 | 0.3438 | No | ||
| 93 | STRN | 3902 | 0.891 | 0.3456 | No | ||
| 94 | NUP153 | 3954 | 0.880 | 0.3458 | No | ||
| 95 | CTBP2 | 3998 | 0.872 | 0.3464 | No | ||
| 96 | MYST4 | 4030 | 0.867 | 0.3475 | No | ||
| 97 | NCK1 | 4115 | 0.853 | 0.3461 | No | ||
| 98 | USP24 | 4142 | 0.848 | 0.3474 | No | ||
| 99 | TIPARP | 4146 | 0.848 | 0.3498 | No | ||
| 100 | AURKA | 4172 | 0.843 | 0.3511 | No | ||
| 101 | ZNF318 | 4234 | 0.834 | 0.3507 | No | ||
| 102 | ATP13A3 | 4402 | 0.803 | 0.3454 | No | ||
| 103 | MICAL2 | 4503 | 0.787 | 0.3431 | No | ||
| 104 | IGF2BP3 | 4555 | 0.777 | 0.3430 | No | ||
| 105 | SP3 | 4662 | 0.763 | 0.3403 | No | ||
| 106 | RBPSUH | 4891 | 0.726 | 0.3320 | No | ||
| 107 | BDKRB2 | 4925 | 0.721 | 0.3326 | No | ||
| 108 | FRYL | 5017 | 0.705 | 0.3304 | No | ||
| 109 | ADORA2B | 5042 | 0.702 | 0.3314 | No | ||
| 110 | HISPPD1 | 5227 | 0.676 | 0.3249 | No | ||
| 111 | RIF1 | 5417 | 0.646 | 0.3181 | No | ||
| 112 | MEGF9 | 5511 | 0.633 | 0.3157 | No | ||
| 113 | ANKRD15 | 5601 | 0.622 | 0.3134 | No | ||
| 114 | CCDC131 | 5739 | 0.601 | 0.3088 | No | ||
| 115 | CTCF | 5879 | 0.578 | 0.3041 | No | ||
| 116 | KIF2A | 5971 | 0.564 | 0.3016 | No | ||
| 117 | PPP1R3C | 6010 | 0.556 | 0.3015 | No | ||
| 118 | CUL1 | 6112 | 0.543 | 0.2984 | No | ||
| 119 | ID1 | 6328 | 0.509 | 0.2900 | No | ||
| 120 | FNBP1L | 6685 | 0.461 | 0.2749 | No | ||
| 121 | MECP2 | 6723 | 0.456 | 0.2746 | No | ||
| 122 | SHOC2 | 6852 | 0.437 | 0.2700 | No | ||
| 123 | E2F3 | 7113 | 0.406 | 0.2592 | No | ||
| 124 | VEGFC | 7469 | 0.357 | 0.2439 | No | ||
| 125 | XPO1 | 7628 | 0.335 | 0.2376 | No | ||
| 126 | UBE2G1 | 7788 | 0.316 | 0.2312 | No | ||
| 127 | UBL3 | 8096 | 0.273 | 0.2178 | No | ||
| 128 | THRAP2 | 8528 | 0.214 | 0.1986 | No | ||
| 129 | ZZZ3 | 8870 | 0.166 | 0.1834 | No | ||
| 130 | BAZ1A | 9115 | 0.131 | 0.1726 | No | ||
| 131 | WDHD1 | 9158 | 0.126 | 0.1710 | No | ||
| 132 | CCNB2 | 9421 | 0.086 | 0.1592 | No | ||
| 133 | KLHL9 | 9669 | 0.048 | 0.1479 | No | ||
| 134 | KIF14 | 10611 | -0.095 | 0.1049 | No | ||
| 135 | GCLC | 10820 | -0.127 | 0.0957 | No | ||
| 136 | ADNP | 10963 | -0.150 | 0.0896 | No | ||
| 137 | TAF5L | 11073 | -0.165 | 0.0850 | No | ||
| 138 | MLH3 | 11090 | -0.167 | 0.0848 | No | ||
| 139 | IRS1 | 11122 | -0.170 | 0.0839 | No | ||
| 140 | PRSS12 | 11420 | -0.207 | 0.0708 | No | ||
| 141 | ZBED4 | 11650 | -0.234 | 0.0609 | No | ||
| 142 | NAV3 | 11788 | -0.248 | 0.0554 | No | ||
| 143 | NFE2L2 | 12111 | -0.285 | 0.0414 | No | ||
| 144 | WEE1 | 12721 | -0.358 | 0.0144 | No | ||
| 145 | CCNE1 | 12750 | -0.362 | 0.0141 | No | ||
| 146 | MTMR6 | 13417 | -0.441 | -0.0152 | No | ||
| 147 | PER2 | 13775 | -0.483 | -0.0303 | No | ||
| 148 | FGFR2 | 13801 | -0.486 | -0.0300 | No | ||
| 149 | BIRC2 | 14071 | -0.516 | -0.0409 | No | ||
| 150 | AGPAT7 | 14124 | -0.523 | -0.0417 | No | ||
| 151 | PIK3C3 | 14267 | -0.541 | -0.0467 | No | ||
| 152 | SMAD5 | 14493 | -0.569 | -0.0554 | No | ||
| 153 | LPHN2 | 14494 | -0.569 | -0.0537 | No | ||
| 154 | PHLPP | 14572 | -0.578 | -0.0556 | No | ||
| 155 | AJAP1 | 14705 | -0.597 | -0.0599 | No | ||
| 156 | TBC1D8 | 14761 | -0.602 | -0.0607 | No | ||
| 157 | DTX2 | 15022 | -0.630 | -0.0708 | No | ||
| 158 | BTBD3 | 15106 | -0.640 | -0.0727 | No | ||
| 159 | AHDC1 | 15708 | -0.716 | -0.0983 | No | ||
| 160 | TUBGCP3 | 15749 | -0.721 | -0.0981 | No | ||
| 161 | KBTBD2 | 16017 | -0.760 | -0.1081 | No | ||
| 162 | CASP8 | 16284 | -0.795 | -0.1180 | No | ||
| 163 | NAGK | 16497 | -0.822 | -0.1254 | No | ||
| 164 | EIF2C2 | 16569 | -0.831 | -0.1262 | No | ||
| 165 | DYRK1A | 16663 | -0.844 | -0.1280 | No | ||
| 166 | RAPGEF2 | 16674 | -0.845 | -0.1260 | No | ||
| 167 | PLEKHC1 | 16700 | -0.847 | -0.1247 | No | ||
| 168 | RCHY1 | 16909 | -0.877 | -0.1317 | No | ||
| 169 | SPEN | 17131 | -0.912 | -0.1392 | No | ||
| 170 | BCL7A | 17212 | -0.925 | -0.1402 | No | ||
| 171 | BCAR3 | 17447 | -0.961 | -0.1481 | No | ||
| 172 | POLE2 | 17509 | -0.973 | -0.1481 | No | ||
| 173 | HDHD1A | 17649 | -0.998 | -0.1516 | No | ||
| 174 | SERTAD2 | 17686 | -1.005 | -0.1503 | No | ||
| 175 | TTC9 | 18035 | -1.064 | -0.1632 | No | ||
| 176 | DOCK4 | 18046 | -1.067 | -0.1605 | No | ||
| 177 | OTUD4 | 18064 | -1.070 | -0.1582 | No | ||
| 178 | MORC3 | 18087 | -1.074 | -0.1561 | No | ||
| 179 | SART3 | 18091 | -1.074 | -0.1530 | No | ||
| 180 | MEIS1 | 18116 | -1.078 | -0.1510 | No | ||
| 181 | FOXJ3 | 18123 | -1.079 | -0.1481 | No | ||
| 182 | CDKN1B | 18144 | -1.082 | -0.1459 | No | ||
| 183 | WWC1 | 18151 | -1.084 | -0.1430 | No | ||
| 184 | KEAP1 | 18484 | -1.152 | -0.1549 | No | ||
| 185 | ADCY9 | 18500 | -1.154 | -0.1522 | No | ||
| 186 | E2F5 | 18509 | -1.155 | -0.1492 | No | ||
| 187 | MTUS1 | 18631 | -1.180 | -0.1513 | No | ||
| 188 | FEM1B | 18671 | -1.188 | -0.1496 | No | ||
| 189 | ZCCHC14 | 19075 | -1.276 | -0.1644 | No | ||
| 190 | CREB3L2 | 19177 | -1.305 | -0.1653 | No | ||
| 191 | CDYL | 19354 | -1.355 | -0.1694 | No | ||
| 192 | LSM14A | 19398 | -1.365 | -0.1674 | No | ||
| 193 | NPAS2 | 19533 | -1.405 | -0.1694 | No | ||
| 194 | PTK2 | 19751 | -1.476 | -0.1751 | No | ||
| 195 | XPA | 19834 | -1.507 | -0.1745 | No | ||
| 196 | MYC | 19912 | -1.534 | -0.1735 | No | ||
| 197 | DOCK9 | 20059 | -1.593 | -0.1756 | No | ||
| 198 | FYN | 20107 | -1.612 | -0.1730 | No | ||
| 199 | INSIG2 | 20274 | -1.686 | -0.1757 | No | ||
| 200 | CCDC93 | 20371 | -1.731 | -0.1751 | No | ||
| 201 | PRKCD | 20514 | -1.805 | -0.1763 | No | ||
| 202 | DDIT4 | 20578 | -1.837 | -0.1738 | No | ||
| 203 | ASXL1 | 20608 | -1.851 | -0.1698 | No | ||
| 204 | ADRB2 | 20660 | -1.884 | -0.1666 | No | ||
| 205 | MN1 | 20768 | -1.946 | -0.1658 | No | ||
| 206 | PPRC1 | 20776 | -1.955 | -0.1604 | No | ||
| 207 | POGZ | 20782 | -1.958 | -0.1549 | No | ||
| 208 | SKAP2 | 20913 | -2.044 | -0.1549 | No | ||
| 209 | NFKB1 | 20944 | -2.065 | -0.1502 | No | ||
| 210 | ARID5B | 20973 | -2.086 | -0.1454 | No | ||
| 211 | NR2F2 | 21012 | -2.123 | -0.1410 | No | ||
| 212 | PRKCBP1 | 21031 | -2.135 | -0.1355 | No | ||
| 213 | GNE | 21103 | -2.197 | -0.1324 | No | ||
| 214 | SLC7A1 | 21110 | -2.202 | -0.1262 | No | ||
| 215 | ARHGEF7 | 21179 | -2.271 | -0.1227 | No | ||
| 216 | MAST4 | 21309 | -2.433 | -0.1215 | No | ||
| 217 | AYTL2 | 21312 | -2.435 | -0.1145 | No | ||
| 218 | PIK3R4 | 21313 | -2.435 | -0.1073 | No | ||
| 219 | RAI17 | 21316 | -2.439 | -0.1003 | No | ||
| 220 | SH2B3 | 21318 | -2.442 | -0.0932 | No | ||
| 221 | FOXO1A | 21342 | -2.471 | -0.0870 | No | ||
| 222 | SLC9A6 | 21433 | -2.647 | -0.0834 | No | ||
| 223 | CBLB | 21453 | -2.694 | -0.0764 | No | ||
| 224 | ZBTB24 | 21497 | -2.779 | -0.0702 | No | ||
| 225 | SMAD3 | 21501 | -2.785 | -0.0622 | No | ||
| 226 | PIP5K2B | 21508 | -2.815 | -0.0542 | No | ||
| 227 | PSCD1 | 21562 | -2.950 | -0.0480 | No | ||
| 228 | RRS1 | 21621 | -3.150 | -0.0415 | No | ||
| 229 | ZHX2 | 21654 | -3.267 | -0.0334 | No | ||
| 230 | TLE4 | 21695 | -3.506 | -0.0249 | No | ||
| 231 | POLS | 21723 | -3.647 | -0.0155 | No | ||
| 232 | NCOA3 | 21812 | -4.359 | -0.0068 | No | ||
| 233 | SMAD7 | 21818 | -4.423 | 0.0059 | No |