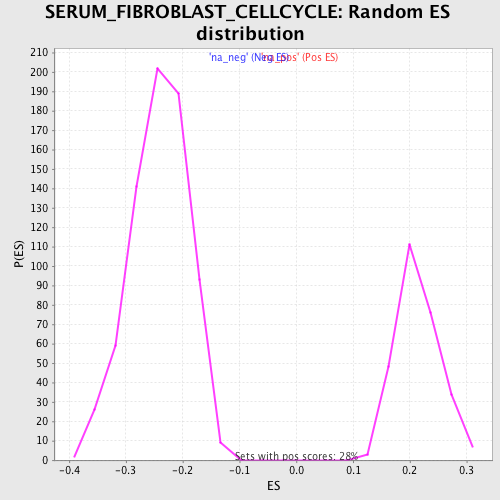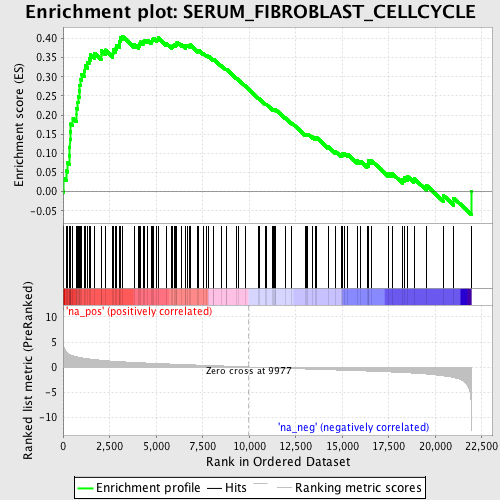
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set02_ATM_minus_versus_BT_ATM_minus |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_pos |
| GeneSet | SERUM_FIBROBLAST_CELLCYCLE |
| Enrichment Score (ES) | 0.40632948 |
| Normalized Enrichment Score (NES) | 1.9057684 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.033782907 |
| FWER p-Value | 0.561 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | SMC4 | 34 | 4.099 | 0.0345 | Yes | ||
| 2 | MPHOSPH1 | 167 | 3.011 | 0.0550 | Yes | ||
| 3 | RAD51AP1 | 254 | 2.713 | 0.0750 | Yes | ||
| 4 | HMMR | 338 | 2.527 | 0.0934 | Yes | ||
| 5 | ABCC5 | 348 | 2.519 | 0.1152 | Yes | ||
| 6 | ATAD2 | 369 | 2.483 | 0.1361 | Yes | ||
| 7 | FBXL20 | 393 | 2.432 | 0.1565 | Yes | ||
| 8 | CASP3 | 413 | 2.406 | 0.1768 | Yes | ||
| 9 | TYMS | 530 | 2.236 | 0.1912 | Yes | ||
| 10 | KIF23 | 717 | 2.037 | 0.2006 | Yes | ||
| 11 | MLF1IP | 719 | 2.033 | 0.2184 | Yes | ||
| 12 | HELLS | 786 | 1.977 | 0.2328 | Yes | ||
| 13 | USP1 | 805 | 1.961 | 0.2493 | Yes | ||
| 14 | CENPF | 863 | 1.909 | 0.2635 | Yes | ||
| 15 | PSRC1 | 897 | 1.886 | 0.2785 | Yes | ||
| 16 | CENPA | 937 | 1.859 | 0.2931 | Yes | ||
| 17 | DHFR | 983 | 1.831 | 0.3072 | Yes | ||
| 18 | TOP2A | 1137 | 1.740 | 0.3155 | Yes | ||
| 19 | PTTG1 | 1175 | 1.721 | 0.3290 | Yes | ||
| 20 | ESCO2 | 1315 | 1.647 | 0.3371 | Yes | ||
| 21 | UHRF1 | 1395 | 1.605 | 0.3476 | Yes | ||
| 22 | ANKRD10 | 1482 | 1.569 | 0.3575 | Yes | ||
| 23 | FOXM1 | 1672 | 1.493 | 0.3620 | Yes | ||
| 24 | MCM8 | 2040 | 1.349 | 0.3570 | Yes | ||
| 25 | CCDC99 | 2042 | 1.348 | 0.3689 | Yes | ||
| 26 | BUB1 | 2260 | 1.274 | 0.3701 | Yes | ||
| 27 | SDC1 | 2679 | 1.157 | 0.3612 | Yes | ||
| 28 | EZH2 | 2683 | 1.156 | 0.3712 | Yes | ||
| 29 | TRIP13 | 2835 | 1.121 | 0.3742 | Yes | ||
| 30 | PHTF2 | 2885 | 1.109 | 0.3817 | Yes | ||
| 31 | TPX2 | 3023 | 1.077 | 0.3849 | Yes | ||
| 32 | UBE2C | 3043 | 1.072 | 0.3935 | Yes | ||
| 33 | GAS2L3 | 3064 | 1.068 | 0.4020 | Yes | ||
| 34 | CDCA8 | 3170 | 1.043 | 0.4063 | Yes | ||
| 35 | SPAG5 | 3843 | 0.902 | 0.3835 | No | ||
| 36 | CKS1B | 4061 | 0.861 | 0.3811 | No | ||
| 37 | KNTC1 | 4123 | 0.851 | 0.3858 | No | ||
| 38 | AURKA | 4172 | 0.843 | 0.3911 | No | ||
| 39 | GINS3 | 4297 | 0.822 | 0.3926 | No | ||
| 40 | CDCA5 | 4390 | 0.805 | 0.3955 | No | ||
| 41 | DLG7 | 4543 | 0.779 | 0.3954 | No | ||
| 42 | MCM5 | 4731 | 0.752 | 0.3934 | No | ||
| 43 | CKAP2 | 4778 | 0.744 | 0.3979 | No | ||
| 44 | ANP32E | 4873 | 0.729 | 0.4000 | No | ||
| 45 | PLK1 | 5036 | 0.703 | 0.3988 | No | ||
| 46 | RRM1 | 5103 | 0.695 | 0.4019 | No | ||
| 47 | PRIM2A | 5542 | 0.628 | 0.3873 | No | ||
| 48 | KIFC1 | 5817 | 0.588 | 0.3800 | No | ||
| 49 | EXO1 | 5902 | 0.574 | 0.3812 | No | ||
| 50 | FEN1 | 5963 | 0.565 | 0.3834 | No | ||
| 51 | CKAP2L | 6047 | 0.552 | 0.3845 | No | ||
| 52 | PRIM1 | 6109 | 0.544 | 0.3865 | No | ||
| 53 | CCNA2 | 6116 | 0.542 | 0.3910 | No | ||
| 54 | WSB1 | 6377 | 0.502 | 0.3835 | No | ||
| 55 | RFC2 | 6568 | 0.478 | 0.3790 | No | ||
| 56 | GTSE1 | 6583 | 0.476 | 0.3825 | No | ||
| 57 | TUBB2C | 6667 | 0.464 | 0.3828 | No | ||
| 58 | MCM6 | 6780 | 0.449 | 0.3816 | No | ||
| 59 | HN1 | 6820 | 0.443 | 0.3837 | No | ||
| 60 | MCM4 | 7237 | 0.389 | 0.3681 | No | ||
| 61 | ADAMTS1 | 7300 | 0.380 | 0.3686 | No | ||
| 62 | LYAR | 7531 | 0.348 | 0.3612 | No | ||
| 63 | CDCA1 | 7707 | 0.325 | 0.3560 | No | ||
| 64 | ANLN | 7835 | 0.309 | 0.3529 | No | ||
| 65 | WDR51A | 8073 | 0.277 | 0.3445 | No | ||
| 66 | AMD1 | 8106 | 0.272 | 0.3454 | No | ||
| 67 | ASF1B | 8507 | 0.217 | 0.3290 | No | ||
| 68 | MAD2L1 | 8752 | 0.183 | 0.3195 | No | ||
| 69 | RFC4 | 8786 | 0.177 | 0.3195 | No | ||
| 70 | TIPIN | 9320 | 0.102 | 0.2960 | No | ||
| 71 | CCNB2 | 9421 | 0.086 | 0.2922 | No | ||
| 72 | PCNA | 9779 | 0.031 | 0.2761 | No | ||
| 73 | LMNB1 | 10486 | -0.076 | 0.2444 | No | ||
| 74 | DONSON | 10527 | -0.083 | 0.2433 | No | ||
| 75 | TIMP1 | 10890 | -0.139 | 0.2280 | No | ||
| 76 | CDC25C | 10914 | -0.143 | 0.2282 | No | ||
| 77 | CCNF | 11263 | -0.189 | 0.2139 | No | ||
| 78 | CKS2 | 11292 | -0.192 | 0.2143 | No | ||
| 79 | EFHC1 | 11359 | -0.201 | 0.2131 | No | ||
| 80 | NCAPH | 11415 | -0.206 | 0.2124 | No | ||
| 81 | MAPK13 | 11422 | -0.207 | 0.2139 | No | ||
| 82 | RECQL4 | 11935 | -0.266 | 0.1928 | No | ||
| 83 | TUBB | 12290 | -0.308 | 0.1793 | No | ||
| 84 | RRM2 | 13015 | -0.393 | 0.1496 | No | ||
| 85 | H2AFX | 13090 | -0.401 | 0.1498 | No | ||
| 86 | MND1 | 13154 | -0.408 | 0.1505 | No | ||
| 87 | MELK | 13403 | -0.439 | 0.1430 | No | ||
| 88 | MET | 13580 | -0.458 | 0.1390 | No | ||
| 89 | PBK | 13590 | -0.459 | 0.1426 | No | ||
| 90 | CDCA7 | 14245 | -0.538 | 0.1174 | No | ||
| 91 | UBE2T | 14636 | -0.587 | 0.1047 | No | ||
| 92 | GMNN | 14935 | -0.622 | 0.0965 | No | ||
| 93 | TACC3 | 15012 | -0.629 | 0.0986 | No | ||
| 94 | FANCG | 15114 | -0.641 | 0.0996 | No | ||
| 95 | PAQR4 | 15290 | -0.663 | 0.0974 | No | ||
| 96 | RANGAP1 | 15793 | -0.727 | 0.0808 | No | ||
| 97 | PWP1 | 15976 | -0.754 | 0.0791 | No | ||
| 98 | BARD1 | 16329 | -0.801 | 0.0701 | No | ||
| 99 | ILF2 | 16411 | -0.811 | 0.0735 | No | ||
| 100 | CDC6 | 16416 | -0.812 | 0.0805 | No | ||
| 101 | FANCA | 16573 | -0.832 | 0.0806 | No | ||
| 102 | SGCD | 17489 | -0.970 | 0.0473 | No | ||
| 103 | YWHAH | 17687 | -1.006 | 0.0471 | No | ||
| 104 | DEPDC1B | 18236 | -1.100 | 0.0317 | No | ||
| 105 | CTNNA1 | 18331 | -1.120 | 0.0373 | No | ||
| 106 | CIITA | 18497 | -1.154 | 0.0399 | No | ||
| 107 | CDC25A | 18854 | -1.230 | 0.0344 | No | ||
| 108 | CDKN3 | 19520 | -1.399 | 0.0163 | No | ||
| 109 | MBOAT1 | 20439 | -1.760 | -0.0103 | No | ||
| 110 | TUBA1 | 20994 | -2.113 | -0.0171 | No | ||
| 111 | ITGB3 | 21928 | -6.890 | 0.0009 | No |