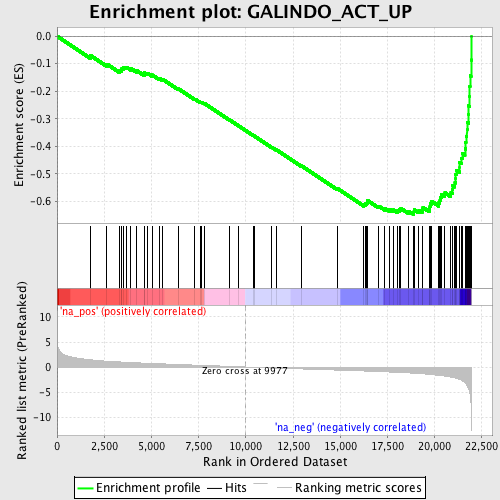
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set02_ATM_minus_versus_BT_ATM_minus |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_neg |
| GeneSet | GALINDO_ACT_UP |
| Enrichment Score (ES) | -0.64732933 |
| Normalized Enrichment Score (NES) | -2.5587301 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.0 |
| FWER p-Value | 0.0 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | PTPRE | 1771 | 1.454 | -0.0704 | No | ||
| 2 | PHLDA1 | 2639 | 1.166 | -0.1015 | No | ||
| 3 | RREB1 | 3299 | 1.013 | -0.1242 | No | ||
| 4 | CCL3 | 3426 | 0.986 | -0.1228 | No | ||
| 5 | SOCS3 | 3433 | 0.984 | -0.1159 | No | ||
| 6 | IER3 | 3521 | 0.965 | -0.1128 | No | ||
| 7 | CEBPB | 3658 | 0.937 | -0.1122 | No | ||
| 8 | PIM1 | 3908 | 0.888 | -0.1171 | No | ||
| 9 | TMEM49 | 4207 | 0.838 | -0.1246 | No | ||
| 10 | VEGF | 4635 | 0.767 | -0.1385 | No | ||
| 11 | CCL5 | 4636 | 0.767 | -0.1329 | No | ||
| 12 | IL18RAP | 4809 | 0.737 | -0.1354 | No | ||
| 13 | ADORA2B | 5042 | 0.702 | -0.1408 | No | ||
| 14 | SLFN4 | 5415 | 0.646 | -0.1531 | No | ||
| 15 | SLC11A1 | 5596 | 0.623 | -0.1568 | No | ||
| 16 | PDGFA | 6424 | 0.496 | -0.1910 | No | ||
| 17 | MARCKSL1 | 7299 | 0.380 | -0.2282 | No | ||
| 18 | CDC42EP4 | 7608 | 0.339 | -0.2398 | No | ||
| 19 | CCL4 | 7646 | 0.333 | -0.2391 | No | ||
| 20 | CSF3 | 7806 | 0.314 | -0.2440 | No | ||
| 21 | TXNIP | 9156 | 0.126 | -0.3048 | No | ||
| 22 | SPP1 | 9619 | 0.056 | -0.3255 | No | ||
| 23 | IRG1 | 10415 | -0.067 | -0.3614 | No | ||
| 24 | GADD45A | 10450 | -0.072 | -0.3624 | No | ||
| 25 | PHLDB1 | 11376 | -0.203 | -0.4032 | No | ||
| 26 | CD14 | 11625 | -0.231 | -0.4129 | No | ||
| 27 | BCL6B | 12936 | -0.383 | -0.4700 | No | ||
| 28 | IL1B | 14865 | -0.614 | -0.5536 | No | ||
| 29 | EHD1 | 16254 | -0.790 | -0.6113 | No | ||
| 30 | FOSL1 | 16325 | -0.800 | -0.6087 | No | ||
| 31 | NFKBIA | 16391 | -0.809 | -0.6058 | No | ||
| 32 | PCDH7 | 16430 | -0.813 | -0.6015 | No | ||
| 33 | HERPUD1 | 16452 | -0.817 | -0.5965 | No | ||
| 34 | PTGS2 | 17046 | -0.900 | -0.6171 | No | ||
| 35 | RHOB | 17370 | -0.948 | -0.6249 | No | ||
| 36 | CXCL2 | 17607 | -0.991 | -0.6285 | No | ||
| 37 | CCL9 | 17821 | -1.025 | -0.6307 | No | ||
| 38 | IL1RN | 18058 | -1.069 | -0.6337 | No | ||
| 39 | PDGFB | 18137 | -1.080 | -0.6294 | No | ||
| 40 | BCL2L11 | 18216 | -1.096 | -0.6249 | No | ||
| 41 | PDLIM7 | 18630 | -1.180 | -0.6352 | No | ||
| 42 | RAC2 | 18896 | -1.240 | -0.6383 | Yes | ||
| 43 | UBC | 18911 | -1.243 | -0.6298 | Yes | ||
| 44 | SCD | 19171 | -1.303 | -0.6322 | Yes | ||
| 45 | TNF | 19341 | -1.351 | -0.6300 | Yes | ||
| 46 | TNIP1 | 19385 | -1.362 | -0.6220 | Yes | ||
| 47 | TNFAIP3 | 19726 | -1.471 | -0.6268 | Yes | ||
| 48 | CCRN4L | 19756 | -1.477 | -0.6174 | Yes | ||
| 49 | SQSTM1 | 19798 | -1.497 | -0.6083 | Yes | ||
| 50 | SRPR | 19826 | -1.506 | -0.5986 | Yes | ||
| 51 | ETS2 | 20232 | -1.668 | -0.6049 | Yes | ||
| 52 | PKM2 | 20286 | -1.691 | -0.5950 | Yes | ||
| 53 | ITGA5 | 20321 | -1.707 | -0.5840 | Yes | ||
| 54 | TNFSF9 | 20359 | -1.724 | -0.5731 | Yes | ||
| 55 | ERRFI1 | 20552 | -1.823 | -0.5686 | Yes | ||
| 56 | ACTG1 | 20869 | -2.009 | -0.5684 | Yes | ||
| 57 | NFKB1 | 20944 | -2.065 | -0.5567 | Yes | ||
| 58 | TNFAIP2 | 20965 | -2.077 | -0.5424 | Yes | ||
| 59 | PLAUR | 21082 | -2.178 | -0.5318 | Yes | ||
| 60 | ICAM1 | 21114 | -2.204 | -0.5171 | Yes | ||
| 61 | DUSP1 | 21121 | -2.210 | -0.5013 | Yes | ||
| 62 | BCL10 | 21159 | -2.247 | -0.4865 | Yes | ||
| 63 | BHLHB2 | 21329 | -2.459 | -0.4763 | Yes | ||
| 64 | CD44 | 21333 | -2.464 | -0.4584 | Yes | ||
| 65 | JUNB | 21440 | -2.670 | -0.4438 | Yes | ||
| 66 | TAPBP | 21470 | -2.730 | -0.4252 | Yes | ||
| 67 | NFKB2 | 21615 | -3.137 | -0.4088 | Yes | ||
| 68 | GLRX | 21641 | -3.213 | -0.3865 | Yes | ||
| 69 | SLFN2 | 21669 | -3.350 | -0.3633 | Yes | ||
| 70 | TRAF1 | 21720 | -3.630 | -0.3390 | Yes | ||
| 71 | DUSP2 | 21727 | -3.672 | -0.3125 | Yes | ||
| 72 | GADD45B | 21806 | -4.294 | -0.2847 | Yes | ||
| 73 | IKBKE | 21814 | -4.363 | -0.2531 | Yes | ||
| 74 | BCL3 | 21865 | -4.977 | -0.2191 | Yes | ||
| 75 | CD83 | 21872 | -5.185 | -0.1815 | Yes | ||
| 76 | TSC22D1 | 21882 | -5.376 | -0.1426 | Yes | ||
| 77 | NAB2 | 21936 | -8.017 | -0.0865 | Yes | ||
| 78 | EGR2 | 21944 | -11.902 | 0.0001 | Yes |