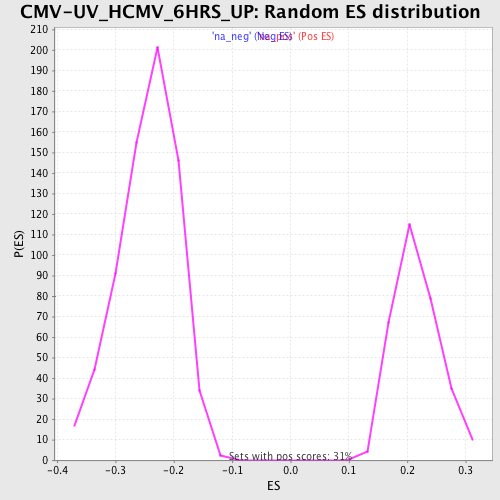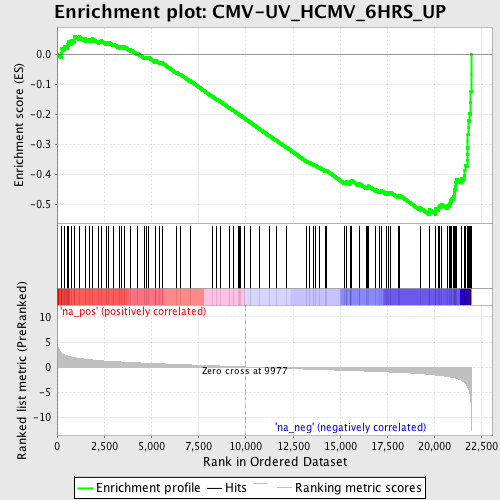
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set02_ATM_minus_versus_BT_ATM_minus |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_neg |
| GeneSet | CMV-UV_HCMV_6HRS_UP |
| Enrichment Score (ES) | -0.5339368 |
| Normalized Enrichment Score (NES) | -2.1811314 |
| Nominal p-value | 0.0 |
| FDR q-value | 8.87547E-4 |
| FWER p-Value | 0.0090 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | RABGAP1L | 230 | 2.821 | 0.0055 | No | ||
| 2 | SOCS1 | 249 | 2.736 | 0.0202 | No | ||
| 3 | SOS1 | 370 | 2.483 | 0.0288 | No | ||
| 4 | ELF1 | 551 | 2.205 | 0.0331 | No | ||
| 5 | BTRC | 602 | 2.155 | 0.0431 | No | ||
| 6 | EPHA5 | 772 | 1.991 | 0.0467 | No | ||
| 7 | PLA2G4A | 906 | 1.879 | 0.0512 | No | ||
| 8 | AGTPBP1 | 908 | 1.877 | 0.0619 | No | ||
| 9 | PTTG1 | 1175 | 1.721 | 0.0595 | No | ||
| 10 | KIF5C | 1517 | 1.555 | 0.0527 | No | ||
| 11 | CCRK | 1731 | 1.471 | 0.0513 | No | ||
| 12 | TNFSF10 | 1870 | 1.406 | 0.0530 | No | ||
| 13 | TAC1 | 2214 | 1.291 | 0.0446 | No | ||
| 14 | NR4A2 | 2339 | 1.246 | 0.0460 | No | ||
| 15 | TROVE2 | 2600 | 1.177 | 0.0408 | No | ||
| 16 | AUTS2 | 2733 | 1.143 | 0.0412 | No | ||
| 17 | FGFR3 | 2996 | 1.082 | 0.0354 | No | ||
| 18 | DPT | 3321 | 1.007 | 0.0263 | No | ||
| 19 | CD38 | 3410 | 0.988 | 0.0279 | No | ||
| 20 | RSAD2 | 3570 | 0.954 | 0.0260 | No | ||
| 21 | CPEB3 | 3883 | 0.894 | 0.0168 | No | ||
| 22 | CHEK2 | 4259 | 0.830 | 0.0043 | No | ||
| 23 | CCL5 | 4636 | 0.767 | -0.0085 | No | ||
| 24 | UBE2S | 4749 | 0.749 | -0.0094 | No | ||
| 25 | IL1RAP | 4850 | 0.732 | -0.0098 | No | ||
| 26 | GTF2B | 5200 | 0.680 | -0.0219 | No | ||
| 27 | COL9A3 | 5204 | 0.679 | -0.0182 | No | ||
| 28 | COL15A1 | 5431 | 0.644 | -0.0249 | No | ||
| 29 | TLR3 | 5570 | 0.625 | -0.0277 | No | ||
| 30 | FGF2 | 6333 | 0.508 | -0.0597 | No | ||
| 31 | IL11 | 6517 | 0.484 | -0.0653 | No | ||
| 32 | CCL8 | 7055 | 0.413 | -0.0875 | No | ||
| 33 | CRYGD | 8243 | 0.255 | -0.1404 | No | ||
| 34 | GALNT3 | 8456 | 0.224 | -0.1489 | No | ||
| 35 | MX1 | 8641 | 0.199 | -0.1562 | No | ||
| 36 | BAZ1A | 9115 | 0.131 | -0.1771 | No | ||
| 37 | IGFBP2 | 9330 | 0.101 | -0.1863 | No | ||
| 38 | AREG | 9596 | 0.061 | -0.1981 | No | ||
| 39 | SLC7A5 | 9674 | 0.047 | -0.2013 | No | ||
| 40 | BRCA2 | 9708 | 0.043 | -0.2026 | No | ||
| 41 | DNAJB9 | 9907 | 0.011 | -0.2116 | No | ||
| 42 | TAF13 | 10268 | -0.045 | -0.2278 | No | ||
| 43 | MSX1 | 10717 | -0.110 | -0.2477 | No | ||
| 44 | CH25H | 11232 | -0.186 | -0.2702 | No | ||
| 45 | ADAMTS3 | 11609 | -0.229 | -0.2861 | No | ||
| 46 | NR4A3 | 12167 | -0.291 | -0.3100 | No | ||
| 47 | OAS2 | 13216 | -0.415 | -0.3556 | No | ||
| 48 | CCL19 | 13388 | -0.437 | -0.3609 | No | ||
| 49 | MX2 | 13592 | -0.459 | -0.3676 | No | ||
| 50 | TRIM21 | 13689 | -0.472 | -0.3693 | No | ||
| 51 | HBEGF | 13904 | -0.499 | -0.3763 | No | ||
| 52 | TRADD | 14236 | -0.537 | -0.3884 | No | ||
| 53 | GCH1 | 14251 | -0.539 | -0.3860 | No | ||
| 54 | RQCD1 | 15251 | -0.657 | -0.4280 | No | ||
| 55 | TFEC | 15308 | -0.666 | -0.4268 | No | ||
| 56 | TREX1 | 15352 | -0.672 | -0.4249 | No | ||
| 57 | PDE1A | 15521 | -0.689 | -0.4287 | No | ||
| 58 | RGS6 | 15564 | -0.696 | -0.4267 | No | ||
| 59 | PMAIP1 | 15566 | -0.696 | -0.4228 | No | ||
| 60 | BAMBI | 15624 | -0.705 | -0.4214 | No | ||
| 61 | PPP1R16B | 15998 | -0.757 | -0.4341 | No | ||
| 62 | IL6 | 16016 | -0.760 | -0.4306 | No | ||
| 63 | NFKBIA | 16391 | -0.809 | -0.4431 | No | ||
| 64 | PCDH9 | 16469 | -0.819 | -0.4420 | No | ||
| 65 | GEM | 16500 | -0.823 | -0.4387 | No | ||
| 66 | RIPK1 | 16874 | -0.871 | -0.4508 | No | ||
| 67 | NP | 17099 | -0.908 | -0.4559 | No | ||
| 68 | PIK3R3 | 17183 | -0.920 | -0.4545 | No | ||
| 69 | MYD88 | 17441 | -0.960 | -0.4608 | No | ||
| 70 | ING1 | 17555 | -0.982 | -0.4604 | No | ||
| 71 | CSTF2 | 17662 | -1.000 | -0.4596 | No | ||
| 72 | POLG2 | 18066 | -1.070 | -0.4719 | No | ||
| 73 | ACTL6B | 18155 | -1.085 | -0.4698 | No | ||
| 74 | RBBP6 | 19244 | -1.322 | -0.5121 | No | ||
| 75 | EGFR | 19722 | -1.469 | -0.5256 | Yes | ||
| 76 | TNFAIP3 | 19726 | -1.471 | -0.5174 | Yes | ||
| 77 | CRY1 | 20063 | -1.594 | -0.5237 | Yes | ||
| 78 | ATF3 | 20071 | -1.597 | -0.5149 | Yes | ||
| 79 | ELN | 20216 | -1.660 | -0.5121 | Yes | ||
| 80 | MEF2C | 20238 | -1.670 | -0.5036 | Yes | ||
| 81 | TNFSF9 | 20359 | -1.724 | -0.4993 | Yes | ||
| 82 | CREM | 20689 | -1.903 | -0.5035 | Yes | ||
| 83 | PFKFB2 | 20777 | -1.955 | -0.4964 | Yes | ||
| 84 | IFIT3 | 20830 | -1.985 | -0.4875 | Yes | ||
| 85 | DOPEY2 | 20921 | -2.050 | -0.4800 | Yes | ||
| 86 | TDRD7 | 21023 | -2.131 | -0.4725 | Yes | ||
| 87 | CX3CR1 | 21036 | -2.142 | -0.4609 | Yes | ||
| 88 | STX11 | 21074 | -2.169 | -0.4502 | Yes | ||
| 89 | ICAM1 | 21114 | -2.204 | -0.4395 | Yes | ||
| 90 | INDO | 21127 | -2.217 | -0.4275 | Yes | ||
| 91 | ISG20 | 21175 | -2.270 | -0.4167 | Yes | ||
| 92 | JUNB | 21440 | -2.670 | -0.4136 | Yes | ||
| 93 | PSCD1 | 21562 | -2.950 | -0.4024 | Yes | ||
| 94 | IRF7 | 21611 | -3.117 | -0.3869 | Yes | ||
| 95 | NFKB2 | 21615 | -3.137 | -0.3692 | Yes | ||
| 96 | DUSP2 | 21727 | -3.672 | -0.3534 | Yes | ||
| 97 | ZC3HAV1 | 21738 | -3.744 | -0.3326 | Yes | ||
| 98 | RELB | 21749 | -3.826 | -0.3113 | Yes | ||
| 99 | RRAD | 21770 | -3.995 | -0.2895 | Yes | ||
| 100 | IFI27 | 21771 | -4.003 | -0.2668 | Yes | ||
| 101 | CCND2 | 21792 | -4.179 | -0.2440 | Yes | ||
| 102 | IRF4 | 21795 | -4.216 | -0.2201 | Yes | ||
| 103 | IER2 | 21848 | -4.749 | -0.1955 | Yes | ||
| 104 | NR4A1 | 21923 | -6.633 | -0.1612 | Yes | ||
| 105 | SAMHD1 | 21929 | -6.895 | -0.1223 | Yes | ||
| 106 | EGR3 | 21943 | -9.744 | -0.0675 | Yes | ||
| 107 | EGR2 | 21944 | -11.902 | 0.0001 | Yes |