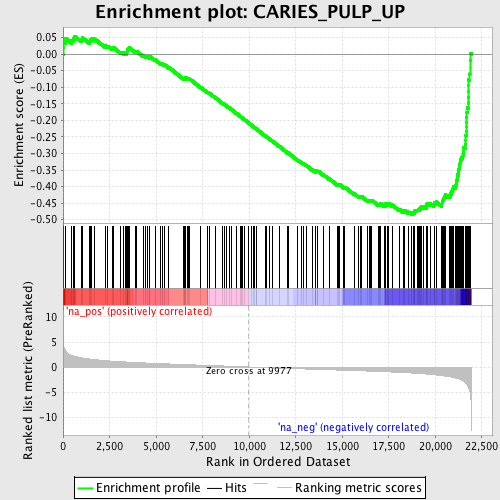
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set02_ATM_minus_versus_BT_ATM_minus |
| Phenotype | NoPhenotypeAvailable |
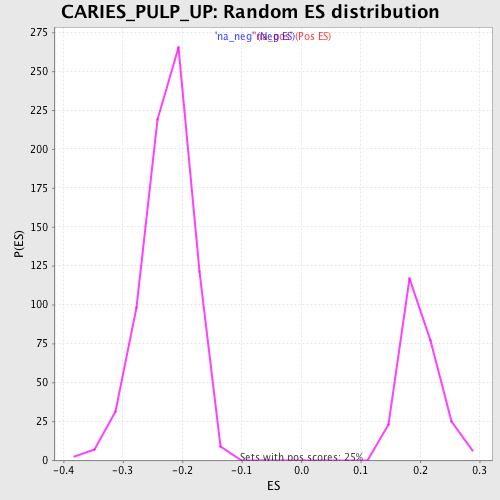
| Upregulated in class | na_neg |
| GeneSet | CARIES_PULP_UP |
| Enrichment Score (ES) | -0.4842219 |
| Normalized Enrichment Score (NES) | -2.156214 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.0010275602 |
| FWER p-Value | 0.013 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | PLAC8 | 12 | 5.058 | 0.0209 | No | ||
| 2 | CD93 | 30 | 4.291 | 0.0382 | No | ||
| 3 | TNFAIP8 | 149 | 3.164 | 0.0462 | No | ||
| 4 | EIF1AY | 450 | 2.349 | 0.0424 | No | ||
| 5 | EVI2A | 537 | 2.229 | 0.0478 | No | ||
| 6 | SLA | 597 | 2.161 | 0.0543 | No | ||
| 7 | VCAM1 | 984 | 1.831 | 0.0443 | No | ||
| 8 | ALOX5AP | 1046 | 1.790 | 0.0491 | No | ||
| 9 | PTPRC | 1419 | 1.597 | 0.0388 | No | ||
| 10 | GZMB | 1463 | 1.577 | 0.0435 | No | ||
| 11 | PECAM1 | 1551 | 1.540 | 0.0460 | No | ||
| 12 | SERPINB2 | 1695 | 1.487 | 0.0457 | No | ||
| 13 | TNFAIP6 | 2256 | 1.275 | 0.0254 | No | ||
| 14 | DDX3Y | 2397 | 1.224 | 0.0241 | No | ||
| 15 | PHLDA1 | 2639 | 1.166 | 0.0180 | No | ||
| 16 | FGL2 | 2725 | 1.145 | 0.0189 | No | ||
| 17 | SLC1A4 | 3109 | 1.058 | 0.0058 | No | ||
| 18 | SLC7A11 | 3237 | 1.028 | 0.0044 | No | ||
| 19 | ADAMDEC1 | 3361 | 0.999 | 0.0029 | No | ||
| 20 | CCL3 | 3426 | 0.986 | 0.0042 | No | ||
| 21 | SOCS3 | 3433 | 0.984 | 0.0080 | No | ||
| 22 | CEBPD | 3462 | 0.977 | 0.0109 | No | ||
| 23 | JARID1D | 3474 | 0.975 | 0.0145 | No | ||
| 24 | IER3 | 3521 | 0.965 | 0.0165 | No | ||
| 25 | NNMT | 3562 | 0.956 | 0.0187 | No | ||
| 26 | ANGPTL2 | 3881 | 0.894 | 0.0079 | No | ||
| 27 | CCR2 | 3950 | 0.880 | 0.0085 | No | ||
| 28 | DARC | 4299 | 0.822 | -0.0040 | No | ||
| 29 | CD84 | 4412 | 0.800 | -0.0058 | No | ||
| 30 | F13A1 | 4515 | 0.784 | -0.0071 | No | ||
| 31 | CCL5 | 4636 | 0.767 | -0.0094 | No | ||
| 32 | PLA2G7 | 4664 | 0.762 | -0.0074 | No | ||
| 33 | PTP4A3 | 4945 | 0.717 | -0.0172 | No | ||
| 34 | CYBB | 5230 | 0.675 | -0.0274 | No | ||
| 35 | ABCA1 | 5343 | 0.657 | -0.0298 | No | ||
| 36 | COL15A1 | 5431 | 0.644 | -0.0311 | No | ||
| 37 | PBEF1 | 5684 | 0.610 | -0.0401 | No | ||
| 38 | CFI | 6473 | 0.490 | -0.0742 | No | ||
| 39 | IL11 | 6517 | 0.484 | -0.0741 | No | ||
| 40 | CLEC7A | 6518 | 0.484 | -0.0720 | No | ||
| 41 | SOD2 | 6573 | 0.477 | -0.0725 | No | ||
| 42 | PLAU | 6575 | 0.477 | -0.0705 | No | ||
| 43 | MS4A1 | 6687 | 0.461 | -0.0737 | No | ||
| 44 | C1QL1 | 6719 | 0.457 | -0.0732 | No | ||
| 45 | CD55 | 6802 | 0.445 | -0.0751 | No | ||
| 46 | TNFRSF21 | 7368 | 0.370 | -0.0994 | No | ||
| 47 | RGS2 | 7747 | 0.321 | -0.1154 | No | ||
| 48 | IBSP | 7759 | 0.320 | -0.1146 | No | ||
| 49 | CXCL12 | 7857 | 0.306 | -0.1177 | No | ||
| 50 | SFRP4 | 8213 | 0.260 | -0.1329 | No | ||
| 51 | CYP1A1 | 8571 | 0.208 | -0.1485 | No | ||
| 52 | STAB1 | 8669 | 0.194 | -0.1521 | No | ||
| 53 | FPR1 | 8797 | 0.175 | -0.1572 | No | ||
| 54 | ENAM | 8926 | 0.158 | -0.1624 | No | ||
| 55 | EVI2B | 9065 | 0.136 | -0.1682 | No | ||
| 56 | SAMSN1 | 9342 | 0.099 | -0.1804 | No | ||
| 57 | MGP | 9525 | 0.071 | -0.1885 | No | ||
| 58 | SERPINE2 | 9569 | 0.065 | -0.1902 | No | ||
| 59 | SPP1 | 9619 | 0.056 | -0.1922 | No | ||
| 60 | RNASE1 | 9751 | 0.036 | -0.1980 | No | ||
| 61 | FYB | 9940 | 0.005 | -0.2067 | No | ||
| 62 | FCER1G | 10142 | -0.025 | -0.2158 | No | ||
| 63 | RAB7L1 | 10214 | -0.036 | -0.2189 | No | ||
| 64 | BLNK | 10309 | -0.050 | -0.2230 | No | ||
| 65 | TYROBP | 10381 | -0.062 | -0.2260 | No | ||
| 66 | NCF2 | 10391 | -0.063 | -0.2261 | No | ||
| 67 | TIMP1 | 10890 | -0.139 | -0.2484 | No | ||
| 68 | REEP1 | 10924 | -0.144 | -0.2493 | No | ||
| 69 | VAMP8 | 11082 | -0.166 | -0.2558 | No | ||
| 70 | ADM | 11254 | -0.188 | -0.2629 | No | ||
| 71 | CD14 | 11625 | -0.231 | -0.2789 | No | ||
| 72 | SSR4 | 12064 | -0.279 | -0.2978 | No | ||
| 73 | C3AR1 | 12084 | -0.281 | -0.2975 | No | ||
| 74 | APOC1 | 12619 | -0.347 | -0.3206 | No | ||
| 75 | CPVL | 12786 | -0.365 | -0.3266 | No | ||
| 76 | CRLF1 | 12902 | -0.380 | -0.3303 | No | ||
| 77 | CXCL5 | 13063 | -0.399 | -0.3360 | No | ||
| 78 | CD48 | 13396 | -0.438 | -0.3494 | No | ||
| 79 | CCR1 | 13552 | -0.456 | -0.3546 | No | ||
| 80 | C5AR1 | 13581 | -0.458 | -0.3539 | No | ||
| 81 | ALOX5 | 13586 | -0.459 | -0.3521 | No | ||
| 82 | KYNU | 13662 | -0.469 | -0.3536 | No | ||
| 83 | PRG1 | 13691 | -0.472 | -0.3529 | No | ||
| 84 | LRRC15 | 13987 | -0.508 | -0.3643 | No | ||
| 85 | C1QB | 14292 | -0.544 | -0.3759 | No | ||
| 86 | SELE | 14755 | -0.601 | -0.3946 | No | ||
| 87 | RAB31 | 14807 | -0.607 | -0.3944 | No | ||
| 88 | IL1B | 14865 | -0.614 | -0.3944 | No | ||
| 89 | MSR1 | 15091 | -0.638 | -0.4020 | No | ||
| 90 | CFD | 15140 | -0.644 | -0.4015 | No | ||
| 91 | LRCH4 | 15642 | -0.707 | -0.4215 | No | ||
| 92 | LY96 | 15869 | -0.738 | -0.4288 | No | ||
| 93 | TREM1 | 15981 | -0.754 | -0.4307 | No | ||
| 94 | IL6 | 16016 | -0.760 | -0.4290 | No | ||
| 95 | CTSC | 16368 | -0.806 | -0.4417 | No | ||
| 96 | HCK | 16436 | -0.815 | -0.4414 | No | ||
| 97 | UBD | 16505 | -0.824 | -0.4410 | No | ||
| 98 | CHL1 | 16583 | -0.833 | -0.4410 | No | ||
| 99 | ECGF1 | 16932 | -0.882 | -0.4533 | No | ||
| 100 | CXCR4 | 17018 | -0.894 | -0.4534 | No | ||
| 101 | PTGS2 | 17046 | -0.900 | -0.4508 | No | ||
| 102 | NFIL3 | 17281 | -0.934 | -0.4576 | No | ||
| 103 | PLEK | 17307 | -0.939 | -0.4548 | No | ||
| 104 | BIRC3 | 17339 | -0.944 | -0.4522 | No | ||
| 105 | IL7R | 17421 | -0.957 | -0.4519 | No | ||
| 106 | LBP | 17462 | -0.966 | -0.4496 | No | ||
| 107 | CXCL13 | 17683 | -1.004 | -0.4555 | No | ||
| 108 | IL1RN | 18058 | -1.069 | -0.4681 | No | ||
| 109 | G0S2 | 18303 | -1.114 | -0.4746 | No | ||
| 110 | CXCL1 | 18358 | -1.125 | -0.4723 | No | ||
| 111 | SLPI | 18560 | -1.165 | -0.4766 | No | ||
| 112 | CYBA | 18726 | -1.199 | -0.4791 | Yes | ||
| 113 | CCL2 | 18809 | -1.219 | -0.4778 | Yes | ||
| 114 | RAC2 | 18896 | -1.240 | -0.4765 | Yes | ||
| 115 | ACSL1 | 18905 | -1.241 | -0.4716 | Yes | ||
| 116 | CSF1R | 19015 | -1.265 | -0.4712 | Yes | ||
| 117 | PTGER4 | 19069 | -1.275 | -0.4683 | Yes | ||
| 118 | WARS | 19170 | -1.303 | -0.4673 | Yes | ||
| 119 | INHBA | 19192 | -1.307 | -0.4628 | Yes | ||
| 120 | MTHFD2 | 19279 | -1.331 | -0.4611 | Yes | ||
| 121 | PSCDBP | 19379 | -1.360 | -0.4599 | Yes | ||
| 122 | IL10RA | 19510 | -1.397 | -0.4599 | Yes | ||
| 123 | LCP1 | 19513 | -1.397 | -0.4541 | Yes | ||
| 124 | ANPEP | 19566 | -1.415 | -0.4505 | Yes | ||
| 125 | TNFAIP3 | 19726 | -1.471 | -0.4516 | Yes | ||
| 126 | HCLS1 | 19929 | -1.540 | -0.4544 | Yes | ||
| 127 | ORM1 | 19962 | -1.550 | -0.4493 | Yes | ||
| 128 | LTB | 20040 | -1.586 | -0.4461 | Yes | ||
| 129 | CAPG | 20348 | -1.716 | -0.4529 | Yes | ||
| 130 | SLAMF8 | 20358 | -1.724 | -0.4460 | Yes | ||
| 131 | LCP2 | 20409 | -1.744 | -0.4410 | Yes | ||
| 132 | NCF4 | 20437 | -1.760 | -0.4348 | Yes | ||
| 133 | ARPC1B | 20489 | -1.784 | -0.4295 | Yes | ||
| 134 | UCP2 | 20547 | -1.819 | -0.4245 | Yes | ||
| 135 | LSP1 | 20764 | -1.943 | -0.4262 | Yes | ||
| 136 | THBS4 | 20828 | -1.983 | -0.4207 | Yes | ||
| 137 | CHI3L1 | 20885 | -2.024 | -0.4147 | Yes | ||
| 138 | SKAP2 | 20913 | -2.044 | -0.4073 | Yes | ||
| 139 | CD53 | 20958 | -2.073 | -0.4005 | Yes | ||
| 140 | PLAUR | 21082 | -2.178 | -0.3969 | Yes | ||
| 141 | ICAM1 | 21114 | -2.204 | -0.3890 | Yes | ||
| 142 | GPNMB | 21125 | -2.215 | -0.3801 | Yes | ||
| 143 | MAN2B1 | 21184 | -2.275 | -0.3732 | Yes | ||
| 144 | SORL1 | 21202 | -2.294 | -0.3642 | Yes | ||
| 145 | S100A8 | 21231 | -2.322 | -0.3557 | Yes | ||
| 146 | AQP3 | 21263 | -2.361 | -0.3471 | Yes | ||
| 147 | SAT1 | 21277 | -2.377 | -0.3377 | Yes | ||
| 148 | AIM1 | 21323 | -2.453 | -0.3294 | Yes | ||
| 149 | CD44 | 21333 | -2.464 | -0.3193 | Yes | ||
| 150 | ARHGDIB | 21394 | -2.566 | -0.3112 | Yes | ||
| 151 | TNFRSF7 | 21486 | -2.756 | -0.3038 | Yes | ||
| 152 | LAPTM5 | 21514 | -2.824 | -0.2930 | Yes | ||
| 153 | S100A9 | 21527 | -2.855 | -0.2815 | Yes | ||
| 154 | IRF8 | 21610 | -3.117 | -0.2721 | Yes | ||
| 155 | CD37 | 21635 | -3.190 | -0.2597 | Yes | ||
| 156 | GLRX | 21641 | -3.213 | -0.2463 | Yes | ||
| 157 | APOE | 21663 | -3.318 | -0.2333 | Yes | ||
| 158 | IFI30 | 21674 | -3.379 | -0.2194 | Yes | ||
| 159 | MMP9 | 21679 | -3.409 | -0.2052 | Yes | ||
| 160 | TRIB1 | 21684 | -3.431 | -0.1908 | Yes | ||
| 161 | CTSB | 21696 | -3.512 | -0.1765 | Yes | ||
| 162 | CD74 | 21718 | -3.626 | -0.1621 | Yes | ||
| 163 | IGFBP4 | 21772 | -4.022 | -0.1475 | Yes | ||
| 164 | CTSH | 21778 | -4.077 | -0.1305 | Yes | ||
| 165 | C3 | 21790 | -4.158 | -0.1134 | Yes | ||
| 166 | IRF4 | 21795 | -4.216 | -0.0957 | Yes | ||
| 167 | PIM2 | 21799 | -4.257 | -0.0779 | Yes | ||
| 168 | TNFRSF1B | 21851 | -4.763 | -0.0601 | Yes | ||
| 169 | BCL3 | 21865 | -4.977 | -0.0396 | Yes | ||
| 170 | POU2AF1 | 21866 | -4.982 | -0.0185 | Yes | ||
| 171 | ITGB2 | 21875 | -5.244 | 0.0033 | Yes |