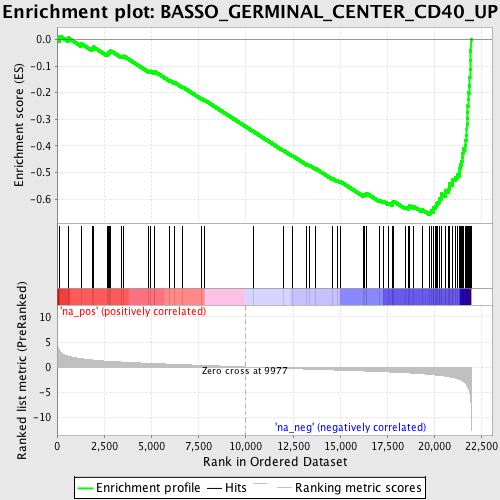
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set02_ATM_minus_versus_BT_ATM_minus |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_neg |
| GeneSet | BASSO_GERMINAL_CENTER_CD40_UP |
| Enrichment Score (ES) | -0.6574811 |
| Normalized Enrichment Score (NES) | -2.660996 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.0 |
| FWER p-Value | 0.0 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | TNFAIP8 | 149 | 3.164 | 0.0122 | No | ||
| 2 | NCKAP1L | 581 | 2.176 | 0.0056 | No | ||
| 3 | NFKBIE | 1265 | 1.671 | -0.0156 | No | ||
| 4 | RGS1 | 1848 | 1.415 | -0.0337 | No | ||
| 5 | HSP90B1 | 1917 | 1.386 | -0.0285 | No | ||
| 6 | PHACTR1 | 2649 | 1.164 | -0.0549 | No | ||
| 7 | SLAMF1 | 2708 | 1.149 | -0.0506 | No | ||
| 8 | ICOSLG | 2782 | 1.132 | -0.0472 | No | ||
| 9 | CHD1 | 2850 | 1.117 | -0.0435 | No | ||
| 10 | CCL3 | 3426 | 0.986 | -0.0639 | No | ||
| 11 | IER3 | 3521 | 0.965 | -0.0624 | No | ||
| 12 | RAB27A | 4851 | 0.732 | -0.1188 | No | ||
| 13 | PTP4A3 | 4945 | 0.717 | -0.1187 | No | ||
| 14 | ELL2 | 5179 | 0.683 | -0.1253 | No | ||
| 15 | MYB | 5181 | 0.683 | -0.1212 | No | ||
| 16 | TCFL5 | 5962 | 0.565 | -0.1535 | No | ||
| 17 | HTR3A | 6199 | 0.527 | -0.1611 | No | ||
| 18 | LGALS1 | 6663 | 0.464 | -0.1795 | No | ||
| 19 | CCL4 | 7646 | 0.333 | -0.2224 | No | ||
| 20 | LHFPL2 | 7815 | 0.312 | -0.2282 | No | ||
| 21 | NCF2 | 10391 | -0.063 | -0.3456 | No | ||
| 22 | CR2 | 11997 | -0.272 | -0.4174 | No | ||
| 23 | TRIP10 | 12447 | -0.325 | -0.4360 | No | ||
| 24 | MAP3K8 | 13234 | -0.417 | -0.4694 | No | ||
| 25 | TCEB3 | 13368 | -0.435 | -0.4729 | No | ||
| 26 | PRG1 | 13691 | -0.472 | -0.4848 | No | ||
| 27 | RXRA | 14616 | -0.584 | -0.5235 | No | ||
| 28 | IL1B | 14865 | -0.614 | -0.5312 | No | ||
| 29 | DTX2 | 15022 | -0.630 | -0.5345 | No | ||
| 30 | EHD1 | 16254 | -0.790 | -0.5861 | No | ||
| 31 | CFLAR | 16298 | -0.798 | -0.5833 | No | ||
| 32 | MAPKAPK2 | 16390 | -0.809 | -0.5825 | No | ||
| 33 | NFKBIA | 16391 | -0.809 | -0.5777 | No | ||
| 34 | SYNGR2 | 17074 | -0.904 | -0.6034 | No | ||
| 35 | PLEK | 17307 | -0.939 | -0.6084 | No | ||
| 36 | PLXNC1 | 17586 | -0.988 | -0.6152 | No | ||
| 37 | NEF3 | 17770 | -1.018 | -0.6174 | No | ||
| 38 | STAT5A | 17775 | -1.018 | -0.6115 | No | ||
| 39 | TMSB4X | 17826 | -1.026 | -0.6076 | No | ||
| 40 | SNF1LK | 18463 | -1.147 | -0.6298 | No | ||
| 41 | BCL2 | 18625 | -1.179 | -0.6300 | No | ||
| 42 | BLR1 | 18680 | -1.189 | -0.6254 | No | ||
| 43 | RAC2 | 18896 | -1.240 | -0.6277 | No | ||
| 44 | TNF | 19341 | -1.351 | -0.6399 | No | ||
| 45 | TNFAIP3 | 19726 | -1.471 | -0.6486 | Yes | ||
| 46 | UNC119 | 19812 | -1.501 | -0.6435 | Yes | ||
| 47 | ZFP36L1 | 19946 | -1.547 | -0.6403 | Yes | ||
| 48 | KCNN4 | 19958 | -1.550 | -0.6314 | Yes | ||
| 49 | ADAM8 | 20045 | -1.588 | -0.6258 | Yes | ||
| 50 | TAP1 | 20082 | -1.603 | -0.6178 | Yes | ||
| 51 | STAT1 | 20172 | -1.640 | -0.6120 | Yes | ||
| 52 | MEF2C | 20238 | -1.670 | -0.6050 | Yes | ||
| 53 | LITAF | 20282 | -1.690 | -0.5968 | Yes | ||
| 54 | CAPG | 20348 | -1.716 | -0.5894 | Yes | ||
| 55 | TNFSF9 | 20359 | -1.724 | -0.5795 | Yes | ||
| 56 | DDIT4 | 20578 | -1.837 | -0.5784 | Yes | ||
| 57 | TMC6 | 20580 | -1.837 | -0.5674 | Yes | ||
| 58 | IRF5 | 20745 | -1.932 | -0.5633 | Yes | ||
| 59 | LTA | 20785 | -1.958 | -0.5533 | Yes | ||
| 60 | FAIM3 | 20812 | -1.974 | -0.5426 | Yes | ||
| 61 | TNFAIP2 | 20965 | -2.077 | -0.5370 | Yes | ||
| 62 | ARID5B | 20973 | -2.086 | -0.5248 | Yes | ||
| 63 | ICAM1 | 21114 | -2.204 | -0.5180 | Yes | ||
| 64 | PIK3CD | 21200 | -2.293 | -0.5080 | Yes | ||
| 65 | SH2B3 | 21318 | -2.442 | -0.4987 | Yes | ||
| 66 | CD44 | 21333 | -2.464 | -0.4845 | Yes | ||
| 67 | CD40 | 21382 | -2.536 | -0.4715 | Yes | ||
| 68 | JUNB | 21440 | -2.670 | -0.4580 | Yes | ||
| 69 | MTMR4 | 21469 | -2.730 | -0.4428 | Yes | ||
| 70 | BATF | 21502 | -2.787 | -0.4275 | Yes | ||
| 71 | LAPTM5 | 21514 | -2.824 | -0.4111 | Yes | ||
| 72 | NFKB2 | 21615 | -3.137 | -0.3968 | Yes | ||
| 73 | SNN | 21655 | -3.277 | -0.3788 | Yes | ||
| 74 | IFI30 | 21674 | -3.379 | -0.3593 | Yes | ||
| 75 | CD74 | 21718 | -3.626 | -0.3395 | Yes | ||
| 76 | TRAF1 | 21720 | -3.630 | -0.3177 | Yes | ||
| 77 | DUSP2 | 21727 | -3.672 | -0.2958 | Yes | ||
| 78 | STK10 | 21759 | -3.915 | -0.2737 | Yes | ||
| 79 | ARHGEF2 | 21769 | -3.994 | -0.2501 | Yes | ||
| 80 | IRF4 | 21795 | -4.216 | -0.2259 | Yes | ||
| 81 | GADD45B | 21806 | -4.294 | -0.2005 | Yes | ||
| 82 | PTK2B | 21837 | -4.616 | -0.1741 | Yes | ||
| 83 | CD83 | 21872 | -5.185 | -0.1444 | Yes | ||
| 84 | ISGF3G | 21881 | -5.367 | -0.1125 | Yes | ||
| 85 | CCL22 | 21893 | -5.744 | -0.0784 | Yes | ||
| 86 | EBI2 | 21905 | -6.015 | -0.0428 | Yes | ||
| 87 | CCR7 | 21932 | -7.419 | 0.0007 | Yes |