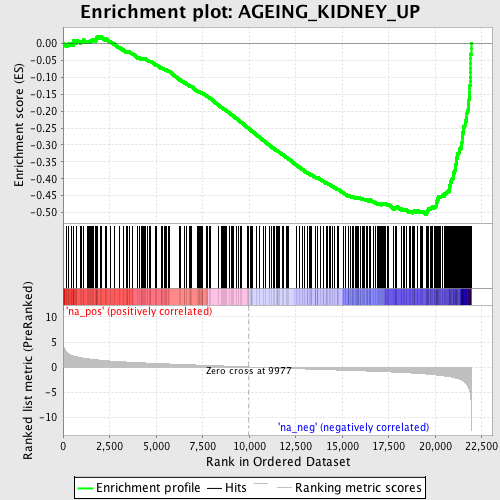
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set02_ATM_minus_versus_BT_ATM_minus |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_neg |
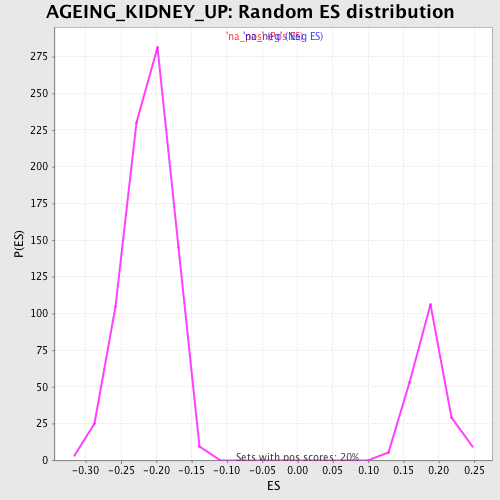
| GeneSet | AGEING_KIDNEY_UP |
| Enrichment Score (ES) | -0.5056518 |
| Normalized Enrichment Score (NES) | -2.394219 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.0 |
| FWER p-Value | 0.0 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | PGM2L1 | 180 | 2.983 | -0.0013 | No | ||
| 2 | APBB1IP | 306 | 2.603 | -0.0010 | No | ||
| 3 | CCND3 | 425 | 2.389 | -0.0008 | No | ||
| 4 | EVI2A | 537 | 2.229 | -0.0007 | No | ||
| 5 | CUGBP2 | 538 | 2.228 | 0.0045 | No | ||
| 6 | RAD54L | 561 | 2.194 | 0.0086 | No | ||
| 7 | NRP1 | 716 | 2.039 | 0.0063 | No | ||
| 8 | PHCA | 737 | 2.021 | 0.0101 | No | ||
| 9 | SLC40A1 | 939 | 1.857 | 0.0052 | No | ||
| 10 | VCAM1 | 984 | 1.831 | 0.0074 | No | ||
| 11 | VWCE | 1068 | 1.776 | 0.0078 | No | ||
| 12 | AATF | 1086 | 1.766 | 0.0111 | No | ||
| 13 | CLIC1 | 1288 | 1.660 | 0.0057 | No | ||
| 14 | CCDC45 | 1344 | 1.634 | 0.0070 | No | ||
| 15 | PTPRC | 1419 | 1.597 | 0.0073 | No | ||
| 16 | DOCK2 | 1485 | 1.568 | 0.0080 | No | ||
| 17 | GZMA | 1534 | 1.548 | 0.0094 | No | ||
| 18 | SLC18A2 | 1574 | 1.532 | 0.0112 | No | ||
| 19 | IFITM1 | 1619 | 1.511 | 0.0127 | No | ||
| 20 | ARHGEF6 | 1736 | 1.469 | 0.0108 | No | ||
| 21 | PTPRE | 1771 | 1.454 | 0.0127 | No | ||
| 22 | CANX | 1790 | 1.442 | 0.0152 | No | ||
| 23 | CD47 | 1791 | 1.442 | 0.0186 | No | ||
| 24 | RGS1 | 1848 | 1.415 | 0.0193 | No | ||
| 25 | PIGR | 1859 | 1.410 | 0.0222 | No | ||
| 26 | ARHGAP15 | 2034 | 1.350 | 0.0173 | No | ||
| 27 | EBF1 | 2045 | 1.346 | 0.0200 | No | ||
| 28 | NT5E | 2290 | 1.264 | 0.0117 | No | ||
| 29 | ARHGAP10 | 2305 | 1.257 | 0.0140 | No | ||
| 30 | GGTA1 | 2535 | 1.189 | 0.0062 | No | ||
| 31 | COL3A1 | 2736 | 1.141 | -0.0004 | No | ||
| 32 | RNF166 | 3053 | 1.070 | -0.0125 | No | ||
| 33 | CDO1 | 3222 | 1.032 | -0.0178 | No | ||
| 34 | CD38 | 3410 | 0.988 | -0.0242 | No | ||
| 35 | SOCS3 | 3433 | 0.984 | -0.0229 | No | ||
| 36 | COL27A1 | 3469 | 0.976 | -0.0222 | No | ||
| 37 | NNMT | 3562 | 0.956 | -0.0242 | No | ||
| 38 | GMFG | 3710 | 0.927 | -0.0288 | No | ||
| 39 | COL1A1 | 4020 | 0.869 | -0.0411 | No | ||
| 40 | PRUNE | 4090 | 0.857 | -0.0422 | No | ||
| 41 | GABRP | 4199 | 0.839 | -0.0453 | No | ||
| 42 | RPS27L | 4200 | 0.839 | -0.0433 | No | ||
| 43 | FER1L3 | 4261 | 0.829 | -0.0441 | No | ||
| 44 | PGLS | 4292 | 0.823 | -0.0436 | No | ||
| 45 | USP31 | 4397 | 0.804 | -0.0465 | No | ||
| 46 | TNFRSF11B | 4403 | 0.803 | -0.0448 | No | ||
| 47 | MMP7 | 4536 | 0.780 | -0.0491 | No | ||
| 48 | CCL5 | 4636 | 0.767 | -0.0519 | No | ||
| 49 | CRIP1 | 4676 | 0.760 | -0.0519 | No | ||
| 50 | MOXD1 | 4708 | 0.755 | -0.0516 | No | ||
| 51 | TUBA3 | 4974 | 0.712 | -0.0622 | No | ||
| 52 | GPR61 | 5028 | 0.704 | -0.0630 | No | ||
| 53 | PIK3CG | 5276 | 0.667 | -0.0728 | No | ||
| 54 | PXDN | 5289 | 0.666 | -0.0718 | No | ||
| 55 | GZMK | 5341 | 0.657 | -0.0726 | No | ||
| 56 | COL15A1 | 5431 | 0.644 | -0.0752 | No | ||
| 57 | RBMS3 | 5501 | 0.634 | -0.0769 | No | ||
| 58 | RAB3GAP1 | 5554 | 0.627 | -0.0779 | No | ||
| 59 | RNF138 | 5644 | 0.615 | -0.0805 | No | ||
| 60 | LIX1 | 5695 | 0.608 | -0.0814 | No | ||
| 61 | GPR56 | 6279 | 0.514 | -0.1071 | No | ||
| 62 | SULF2 | 6323 | 0.509 | -0.1079 | No | ||
| 63 | CLDN3 | 6515 | 0.484 | -0.1156 | No | ||
| 64 | CLEC7A | 6518 | 0.484 | -0.1146 | No | ||
| 65 | SCN5A | 6651 | 0.466 | -0.1196 | No | ||
| 66 | C1S | 6807 | 0.444 | -0.1257 | No | ||
| 67 | C1R | 6837 | 0.440 | -0.1260 | No | ||
| 68 | SPON1 | 6847 | 0.438 | -0.1254 | No | ||
| 69 | CTHRC1 | 6893 | 0.433 | -0.1265 | No | ||
| 70 | FBN1 | 7224 | 0.392 | -0.1408 | No | ||
| 71 | LUM | 7260 | 0.387 | -0.1415 | No | ||
| 72 | ANTXR2 | 7328 | 0.375 | -0.1437 | No | ||
| 73 | COL5A1 | 7329 | 0.375 | -0.1429 | No | ||
| 74 | MGAT1 | 7397 | 0.367 | -0.1451 | No | ||
| 75 | IRAK1 | 7424 | 0.365 | -0.1455 | No | ||
| 76 | VEGFC | 7469 | 0.357 | -0.1466 | No | ||
| 77 | TSPAN1 | 7488 | 0.355 | -0.1466 | No | ||
| 78 | TNC | 7514 | 0.351 | -0.1470 | No | ||
| 79 | DTX3L | 7694 | 0.327 | -0.1545 | No | ||
| 80 | VARS | 7750 | 0.320 | -0.1563 | No | ||
| 81 | NOV | 7853 | 0.306 | -0.1603 | No | ||
| 82 | COL8A2 | 7895 | 0.301 | -0.1615 | No | ||
| 83 | LY75 | 8355 | 0.239 | -0.1821 | No | ||
| 84 | GPX1 | 8530 | 0.214 | -0.1897 | No | ||
| 85 | RARRES1 | 8577 | 0.207 | -0.1913 | No | ||
| 86 | GABRE | 8593 | 0.204 | -0.1915 | No | ||
| 87 | STAB1 | 8669 | 0.194 | -0.1945 | No | ||
| 88 | PROM1 | 8726 | 0.187 | -0.1967 | No | ||
| 89 | RPL28 | 8794 | 0.176 | -0.1994 | No | ||
| 90 | ENAM | 8926 | 0.158 | -0.2050 | No | ||
| 91 | CDH11 | 9049 | 0.140 | -0.2104 | No | ||
| 92 | RNASE6 | 9094 | 0.134 | -0.2121 | No | ||
| 93 | COMP | 9175 | 0.124 | -0.2155 | No | ||
| 94 | SVEP1 | 9304 | 0.104 | -0.2212 | No | ||
| 95 | RAB34 | 9407 | 0.087 | -0.2257 | No | ||
| 96 | LOXL1 | 9507 | 0.074 | -0.2301 | No | ||
| 97 | SERPINE2 | 9569 | 0.065 | -0.2327 | No | ||
| 98 | AMBP | 9930 | 0.006 | -0.2494 | No | ||
| 99 | FYB | 9940 | 0.005 | -0.2498 | No | ||
| 100 | C1QC | 9981 | -0.001 | -0.2516 | No | ||
| 101 | ALPK2 | 10093 | -0.018 | -0.2567 | No | ||
| 102 | FGG | 10114 | -0.022 | -0.2576 | No | ||
| 103 | FCER1G | 10142 | -0.025 | -0.2588 | No | ||
| 104 | STS | 10165 | -0.029 | -0.2597 | No | ||
| 105 | TYROBP | 10381 | -0.062 | -0.2695 | No | ||
| 106 | CASP1 | 10532 | -0.083 | -0.2762 | No | ||
| 107 | CD97 | 10755 | -0.116 | -0.2862 | No | ||
| 108 | TIMP1 | 10890 | -0.139 | -0.2921 | No | ||
| 109 | FAS | 11067 | -0.164 | -0.2999 | No | ||
| 110 | CCDC23 | 11184 | -0.178 | -0.3048 | No | ||
| 111 | CALU | 11279 | -0.191 | -0.3087 | No | ||
| 112 | TBXAS1 | 11377 | -0.203 | -0.3127 | No | ||
| 113 | FDXR | 11474 | -0.214 | -0.3166 | No | ||
| 114 | DOCK11 | 11520 | -0.220 | -0.3182 | No | ||
| 115 | SERPING1 | 11538 | -0.222 | -0.3185 | No | ||
| 116 | APRT | 11562 | -0.224 | -0.3190 | No | ||
| 117 | CD14 | 11625 | -0.231 | -0.3213 | No | ||
| 118 | NUAK1 | 11780 | -0.248 | -0.3279 | No | ||
| 119 | TRIM47 | 11828 | -0.255 | -0.3294 | No | ||
| 120 | TMED3 | 11996 | -0.272 | -0.3365 | No | ||
| 121 | SSR4 | 12064 | -0.279 | -0.3390 | No | ||
| 122 | RPL13A | 12126 | -0.286 | -0.3411 | No | ||
| 123 | RPL35A | 12550 | -0.339 | -0.3599 | No | ||
| 124 | PSMB9 | 12679 | -0.353 | -0.3650 | No | ||
| 125 | TKT | 12867 | -0.374 | -0.3727 | No | ||
| 126 | RPL18 | 12995 | -0.389 | -0.3777 | No | ||
| 127 | GRN | 13105 | -0.403 | -0.3818 | No | ||
| 128 | VASH1 | 13145 | -0.408 | -0.3826 | No | ||
| 129 | MAP3K8 | 13234 | -0.417 | -0.3857 | No | ||
| 130 | SPON2 | 13306 | -0.425 | -0.3880 | No | ||
| 131 | PFN1 | 13354 | -0.432 | -0.3892 | No | ||
| 132 | FN1 | 13360 | -0.434 | -0.3884 | No | ||
| 133 | CCR1 | 13552 | -0.456 | -0.3961 | No | ||
| 134 | SYTL2 | 13556 | -0.456 | -0.3952 | No | ||
| 135 | ALOX5 | 13586 | -0.459 | -0.3955 | No | ||
| 136 | PARVG | 13657 | -0.468 | -0.3976 | No | ||
| 137 | RUNX2 | 13659 | -0.469 | -0.3965 | No | ||
| 138 | PRG1 | 13691 | -0.472 | -0.3969 | No | ||
| 139 | SYMPK | 13814 | -0.488 | -0.4014 | No | ||
| 140 | CCM2 | 13964 | -0.505 | -0.4071 | No | ||
| 141 | KLHDC5 | 13979 | -0.507 | -0.4065 | No | ||
| 142 | VTCN1 | 14131 | -0.523 | -0.4123 | No | ||
| 143 | CFB | 14142 | -0.524 | -0.4115 | No | ||
| 144 | MRAS | 14204 | -0.533 | -0.4131 | No | ||
| 145 | C1QB | 14292 | -0.544 | -0.4158 | No | ||
| 146 | NKG7 | 14349 | -0.551 | -0.4171 | No | ||
| 147 | CORO1A | 14491 | -0.569 | -0.4223 | No | ||
| 148 | EML3 | 14600 | -0.582 | -0.4259 | No | ||
| 149 | REEP4 | 14754 | -0.601 | -0.4316 | No | ||
| 150 | TFPI | 14757 | -0.601 | -0.4303 | No | ||
| 151 | RAB31 | 14807 | -0.607 | -0.4311 | No | ||
| 152 | MSR1 | 15091 | -0.638 | -0.4427 | No | ||
| 153 | BHLHB3 | 15178 | -0.650 | -0.4452 | No | ||
| 154 | COL14A1 | 15309 | -0.666 | -0.4496 | No | ||
| 155 | DOK1 | 15319 | -0.668 | -0.4484 | No | ||
| 156 | TREX1 | 15352 | -0.672 | -0.4484 | No | ||
| 157 | IL17B | 15461 | -0.684 | -0.4517 | No | ||
| 158 | MYO1F | 15552 | -0.694 | -0.4543 | No | ||
| 159 | ANXA2 | 15569 | -0.697 | -0.4534 | No | ||
| 160 | RAB8B | 15628 | -0.706 | -0.4544 | No | ||
| 161 | RPS25 | 15733 | -0.720 | -0.4575 | No | ||
| 162 | ISLR | 15751 | -0.721 | -0.4566 | No | ||
| 163 | FAU | 15816 | -0.730 | -0.4579 | No | ||
| 164 | PSMB8 | 15830 | -0.732 | -0.4567 | No | ||
| 165 | KRTCAP2 | 15846 | -0.734 | -0.4557 | No | ||
| 166 | HTATIP2 | 15880 | -0.739 | -0.4555 | No | ||
| 167 | ACTN1 | 15953 | -0.750 | -0.4571 | No | ||
| 168 | PLOD3 | 15996 | -0.757 | -0.4572 | No | ||
| 169 | EMP3 | 16063 | -0.766 | -0.4585 | No | ||
| 170 | PTPN22 | 16138 | -0.775 | -0.4601 | No | ||
| 171 | CLDN1 | 16184 | -0.782 | -0.4603 | No | ||
| 172 | ANTXR1 | 16279 | -0.795 | -0.4628 | No | ||
| 173 | ST8SIA4 | 16336 | -0.802 | -0.4635 | No | ||
| 174 | TPBG | 16360 | -0.805 | -0.4627 | No | ||
| 175 | RBP1 | 16479 | -0.821 | -0.4662 | No | ||
| 176 | HMHA1 | 16480 | -0.821 | -0.4643 | No | ||
| 177 | PARP12 | 16491 | -0.822 | -0.4628 | No | ||
| 178 | UBD | 16505 | -0.824 | -0.4615 | No | ||
| 179 | WFDC2 | 16702 | -0.848 | -0.4686 | No | ||
| 180 | DOK3 | 16808 | -0.863 | -0.4714 | No | ||
| 181 | RPL35 | 16904 | -0.876 | -0.4737 | No | ||
| 182 | ECGF1 | 16932 | -0.882 | -0.4729 | No | ||
| 183 | SLC12A9 | 17006 | -0.892 | -0.4742 | No | ||
| 184 | SYNGR2 | 17074 | -0.904 | -0.4752 | No | ||
| 185 | AEBP1 | 17129 | -0.912 | -0.4755 | No | ||
| 186 | ASNS | 17144 | -0.915 | -0.4740 | No | ||
| 187 | RTN1 | 17217 | -0.925 | -0.4752 | No | ||
| 188 | STOML1 | 17261 | -0.931 | -0.4750 | No | ||
| 189 | NFKBIZ | 17301 | -0.938 | -0.4746 | No | ||
| 190 | BIRC3 | 17339 | -0.944 | -0.4741 | No | ||
| 191 | CSPG2 | 17454 | -0.963 | -0.4771 | No | ||
| 192 | HGF | 17482 | -0.969 | -0.4761 | No | ||
| 193 | DDX58 | 17778 | -1.019 | -0.4873 | No | ||
| 194 | CHRNA3 | 17833 | -1.027 | -0.4874 | No | ||
| 195 | FRS2 | 17861 | -1.034 | -0.4862 | No | ||
| 196 | PPM1M | 17884 | -1.039 | -0.4848 | No | ||
| 197 | FHOD1 | 17925 | -1.047 | -0.4842 | No | ||
| 198 | TGIF | 17938 | -1.050 | -0.4823 | No | ||
| 199 | DGKQ | 18161 | -1.085 | -0.4900 | No | ||
| 200 | LST1 | 18181 | -1.089 | -0.4883 | No | ||
| 201 | SNCA | 18286 | -1.110 | -0.4905 | No | ||
| 202 | CXCL1 | 18358 | -1.125 | -0.4912 | No | ||
| 203 | VSNL1 | 18433 | -1.141 | -0.4919 | No | ||
| 204 | ARHGAP30 | 18621 | -1.179 | -0.4978 | No | ||
| 205 | FNBP1 | 18676 | -1.189 | -0.4975 | No | ||
| 206 | SLC1A3 | 18760 | -1.208 | -0.4985 | No | ||
| 207 | CCL2 | 18809 | -1.219 | -0.4979 | No | ||
| 208 | CNO | 18880 | -1.237 | -0.4982 | No | ||
| 209 | NFIX | 18888 | -1.239 | -0.4956 | No | ||
| 210 | RAC2 | 18896 | -1.240 | -0.4930 | No | ||
| 211 | CSF1R | 19015 | -1.265 | -0.4955 | No | ||
| 212 | COL1A2 | 19064 | -1.274 | -0.4947 | No | ||
| 213 | MYO5B | 19195 | -1.308 | -0.4977 | No | ||
| 214 | WDR46 | 19260 | -1.327 | -0.4975 | No | ||
| 215 | CARHSP1 | 19320 | -1.346 | -0.4971 | No | ||
| 216 | ADRA2A | 19506 | -1.396 | -0.5024 | Yes | ||
| 217 | IL10RA | 19510 | -1.397 | -0.4992 | Yes | ||
| 218 | DPP3 | 19553 | -1.411 | -0.4979 | Yes | ||
| 219 | RAB33A | 19588 | -1.420 | -0.4961 | Yes | ||
| 220 | TMEM43 | 19589 | -1.421 | -0.4928 | Yes | ||
| 221 | NR2F1 | 19622 | -1.432 | -0.4909 | Yes | ||
| 222 | MALL | 19645 | -1.437 | -0.4885 | Yes | ||
| 223 | IFNAR1 | 19727 | -1.471 | -0.4888 | Yes | ||
| 224 | PAM | 19757 | -1.478 | -0.4867 | Yes | ||
| 225 | HTR7 | 19785 | -1.492 | -0.4845 | Yes | ||
| 226 | MFGE8 | 19837 | -1.509 | -0.4833 | Yes | ||
| 227 | HCLS1 | 19929 | -1.540 | -0.4839 | Yes | ||
| 228 | GGTLA1 | 19973 | -1.555 | -0.4822 | Yes | ||
| 229 | CST6 | 20019 | -1.577 | -0.4806 | Yes | ||
| 230 | LTB | 20040 | -1.586 | -0.4778 | Yes | ||
| 231 | ADAM8 | 20045 | -1.588 | -0.4742 | Yes | ||
| 232 | SLC15A3 | 20046 | -1.589 | -0.4705 | Yes | ||
| 233 | POU4F3 | 20078 | -1.600 | -0.4682 | Yes | ||
| 234 | SLC6A6 | 20094 | -1.607 | -0.4651 | Yes | ||
| 235 | DDEF1 | 20102 | -1.609 | -0.4617 | Yes | ||
| 236 | IKBKB | 20132 | -1.623 | -0.4592 | Yes | ||
| 237 | ATHL1 | 20173 | -1.640 | -0.4572 | Yes | ||
| 238 | TPM3 | 20175 | -1.642 | -0.4534 | Yes | ||
| 239 | TRAFD1 | 20249 | -1.674 | -0.4528 | Yes | ||
| 240 | HCST | 20297 | -1.698 | -0.4510 | Yes | ||
| 241 | SLAMF8 | 20358 | -1.724 | -0.4498 | Yes | ||
| 242 | LCP2 | 20409 | -1.744 | -0.4480 | Yes | ||
| 243 | TPM1 | 20488 | -1.784 | -0.4474 | Yes | ||
| 244 | ARPC1B | 20489 | -1.784 | -0.4432 | Yes | ||
| 245 | CRELD2 | 20569 | -1.832 | -0.4426 | Yes | ||
| 246 | P4HB | 20590 | -1.841 | -0.4392 | Yes | ||
| 247 | SETD7 | 20645 | -1.873 | -0.4373 | Yes | ||
| 248 | PLEKHO1 | 20697 | -1.906 | -0.4352 | Yes | ||
| 249 | PDIA4 | 20753 | -1.937 | -0.4332 | Yes | ||
| 250 | DEGS1 | 20757 | -1.942 | -0.4287 | Yes | ||
| 251 | MMP11 | 20759 | -1.942 | -0.4242 | Yes | ||
| 252 | GNG2 | 20773 | -1.950 | -0.4203 | Yes | ||
| 253 | CD69 | 20802 | -1.970 | -0.4169 | Yes | ||
| 254 | FXYD5 | 20809 | -1.973 | -0.4126 | Yes | ||
| 255 | CXCL14 | 20829 | -1.984 | -0.4088 | Yes | ||
| 256 | FEZ2 | 20858 | -2.002 | -0.4054 | Yes | ||
| 257 | IRAK2 | 20864 | -2.006 | -0.4009 | Yes | ||
| 258 | CORO7 | 20907 | -2.037 | -0.3981 | Yes | ||
| 259 | LTF | 20956 | -2.072 | -0.3954 | Yes | ||
| 260 | CD53 | 20958 | -2.073 | -0.3906 | Yes | ||
| 261 | TNFAIP2 | 20965 | -2.077 | -0.3860 | Yes | ||
| 262 | PARP14 | 20974 | -2.087 | -0.3815 | Yes | ||
| 263 | CX3CR1 | 21036 | -2.142 | -0.3793 | Yes | ||
| 264 | LIG3 | 21049 | -2.151 | -0.3748 | Yes | ||
| 265 | LDLRAP1 | 21072 | -2.163 | -0.3707 | Yes | ||
| 266 | IL10RB | 21090 | -2.188 | -0.3664 | Yes | ||
| 267 | AXL | 21095 | -2.192 | -0.3614 | Yes | ||
| 268 | ARHGAP1 | 21097 | -2.192 | -0.3563 | Yes | ||
| 269 | GPNMB | 21125 | -2.215 | -0.3524 | Yes | ||
| 270 | ITPR3 | 21126 | -2.217 | -0.3472 | Yes | ||
| 271 | MARCH1 | 21141 | -2.228 | -0.3426 | Yes | ||
| 272 | ITM2C | 21158 | -2.247 | -0.3381 | Yes | ||
| 273 | MAN2B1 | 21184 | -2.275 | -0.3339 | Yes | ||
| 274 | PIK3CD | 21200 | -2.293 | -0.3292 | Yes | ||
| 275 | RPN1 | 21204 | -2.295 | -0.3240 | Yes | ||
| 276 | RASSF5 | 21270 | -2.364 | -0.3214 | Yes | ||
| 277 | RPS6KA1 | 21310 | -2.434 | -0.3175 | Yes | ||
| 278 | AYTL2 | 21312 | -2.435 | -0.3118 | Yes | ||
| 279 | RGS19 | 21366 | -2.510 | -0.3084 | Yes | ||
| 280 | ARHGDIB | 21394 | -2.566 | -0.3036 | Yes | ||
| 281 | ARHGAP25 | 21397 | -2.571 | -0.2977 | Yes | ||
| 282 | PRICKLE1 | 21424 | -2.616 | -0.2928 | Yes | ||
| 283 | RIN3 | 21437 | -2.664 | -0.2871 | Yes | ||
| 284 | SAMD9L | 21438 | -2.669 | -0.2808 | Yes | ||
| 285 | SPARCL1 | 21448 | -2.684 | -0.2749 | Yes | ||
| 286 | TCF3 | 21468 | -2.726 | -0.2694 | Yes | ||
| 287 | TAPBP | 21470 | -2.730 | -0.2630 | Yes | ||
| 288 | LAPTM5 | 21514 | -2.824 | -0.2584 | Yes | ||
| 289 | STAT6 | 21519 | -2.830 | -0.2520 | Yes | ||
| 290 | ATP8B2 | 21523 | -2.837 | -0.2454 | Yes | ||
| 291 | ABCC1 | 21592 | -3.079 | -0.2414 | Yes | ||
| 292 | PTPN6 | 21597 | -3.085 | -0.2343 | Yes | ||
| 293 | IRF8 | 21610 | -3.117 | -0.2275 | Yes | ||
| 294 | APOE | 21663 | -3.318 | -0.2222 | Yes | ||
| 295 | CMTM3 | 21694 | -3.502 | -0.2153 | Yes | ||
| 296 | TBC1D2B | 21697 | -3.512 | -0.2072 | Yes | ||
| 297 | CORO1C | 21703 | -3.548 | -0.1991 | Yes | ||
| 298 | STK10 | 21759 | -3.915 | -0.1925 | Yes | ||
| 299 | CCND2 | 21792 | -4.179 | -0.1841 | Yes | ||
| 300 | PSME2 | 21801 | -4.260 | -0.1745 | Yes | ||
| 301 | C9ORF19 | 21807 | -4.299 | -0.1646 | Yes | ||
| 302 | LASP1 | 21823 | -4.444 | -0.1549 | Yes | ||
| 303 | TNFRSF25 | 21833 | -4.603 | -0.1445 | Yes | ||
| 304 | CXCL16 | 21853 | -4.800 | -0.1341 | Yes | ||
| 305 | RUNX3 | 21857 | -4.865 | -0.1229 | Yes | ||
| 306 | ITGB2 | 21875 | -5.244 | -0.1114 | Yes | ||
| 307 | ISGF3G | 21881 | -5.367 | -0.0990 | Yes | ||
| 308 | VIM | 21890 | -5.636 | -0.0861 | Yes | ||
| 309 | TESC | 21895 | -5.748 | -0.0728 | Yes | ||
| 310 | TGFBI | 21900 | -5.856 | -0.0593 | Yes | ||
| 311 | SCOTIN | 21907 | -6.138 | -0.0452 | Yes | ||
| 312 | LYSMD2 | 21911 | -6.208 | -0.0307 | Yes | ||
| 313 | TMSB10 | 21927 | -6.825 | -0.0154 | Yes | ||
| 314 | RGS10 | 21930 | -6.943 | 0.0008 | Yes |