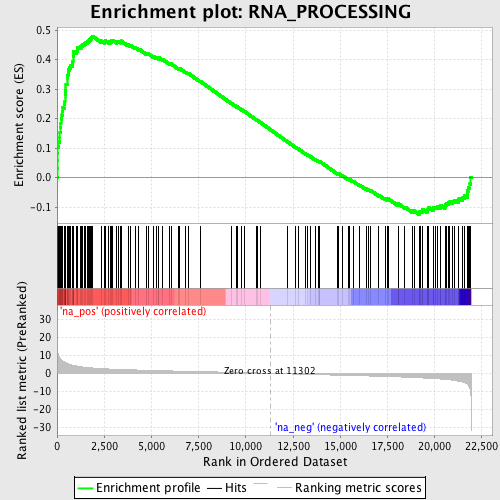
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set01_ATM_minus_versus_SCID |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_pos |
| GeneSet | RNA_PROCESSING |
| Enrichment Score (ES) | 0.47996098 |
| Normalized Enrichment Score (NES) | 2.1532106 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.0019079164 |
| FWER p-Value | 0.012 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | SFRS2 | 12 | 15.098 | 0.0302 | Yes | ||
| 2 | SFRS10 | 19 | 13.401 | 0.0573 | Yes | ||
| 3 | DUSP11 | 26 | 12.581 | 0.0826 | Yes | ||
| 4 | SYNCRIP | 38 | 11.556 | 0.1057 | Yes | ||
| 5 | SFRS11 | 88 | 9.493 | 0.1228 | Yes | ||
| 6 | CRNKL1 | 145 | 8.544 | 0.1376 | Yes | ||
| 7 | SF3A3 | 149 | 8.506 | 0.1548 | Yes | ||
| 8 | PABPC1 | 176 | 8.119 | 0.1702 | Yes | ||
| 9 | DDX20 | 199 | 7.717 | 0.1849 | Yes | ||
| 10 | SFRS7 | 206 | 7.661 | 0.2003 | Yes | ||
| 11 | FUSIP1 | 256 | 7.124 | 0.2125 | Yes | ||
| 12 | SSB | 280 | 6.995 | 0.2257 | Yes | ||
| 13 | CSTF2 | 292 | 6.925 | 0.2394 | Yes | ||
| 14 | NUFIP1 | 393 | 6.197 | 0.2474 | Yes | ||
| 15 | INTS4 | 413 | 6.106 | 0.2590 | Yes | ||
| 16 | CPSF6 | 418 | 6.068 | 0.2712 | Yes | ||
| 17 | PRPF4B | 420 | 6.057 | 0.2835 | Yes | ||
| 18 | SMNDC1 | 444 | 5.924 | 0.2945 | Yes | ||
| 19 | ELAVL4 | 466 | 5.799 | 0.3053 | Yes | ||
| 20 | POP4 | 468 | 5.791 | 0.3171 | Yes | ||
| 21 | HNRPDL | 529 | 5.454 | 0.3255 | Yes | ||
| 22 | NUDT21 | 530 | 5.452 | 0.3366 | Yes | ||
| 23 | SFRS6 | 535 | 5.431 | 0.3475 | Yes | ||
| 24 | NOLC1 | 581 | 5.207 | 0.3560 | Yes | ||
| 25 | IVNS1ABP | 594 | 5.153 | 0.3660 | Yes | ||
| 26 | PRPF3 | 660 | 4.925 | 0.3730 | Yes | ||
| 27 | INTS2 | 730 | 4.716 | 0.3795 | Yes | ||
| 28 | DHX15 | 802 | 4.544 | 0.3855 | Yes | ||
| 29 | RBM6 | 834 | 4.459 | 0.3932 | Yes | ||
| 30 | SFRS1 | 843 | 4.448 | 0.4019 | Yes | ||
| 31 | RNMT | 844 | 4.445 | 0.4109 | Yes | ||
| 32 | SRRM1 | 864 | 4.408 | 0.4190 | Yes | ||
| 33 | INTS12 | 869 | 4.400 | 0.4278 | Yes | ||
| 34 | SNRPA1 | 1035 | 4.018 | 0.4284 | Yes | ||
| 35 | NCBP2 | 1070 | 3.938 | 0.4349 | Yes | ||
| 36 | KIN | 1083 | 3.919 | 0.4424 | Yes | ||
| 37 | RNGTT | 1212 | 3.700 | 0.4440 | Yes | ||
| 38 | TRNT1 | 1312 | 3.537 | 0.4467 | Yes | ||
| 39 | SFRS2IP | 1335 | 3.513 | 0.4528 | Yes | ||
| 40 | INTS8 | 1439 | 3.384 | 0.4550 | Yes | ||
| 41 | CUGBP1 | 1522 | 3.289 | 0.4580 | Yes | ||
| 42 | NOL3 | 1598 | 3.206 | 0.4611 | Yes | ||
| 43 | SFPQ | 1655 | 3.149 | 0.4649 | Yes | ||
| 44 | DGCR8 | 1701 | 3.108 | 0.4692 | Yes | ||
| 45 | PPIG | 1764 | 3.057 | 0.4726 | Yes | ||
| 46 | GRSF1 | 1821 | 3.003 | 0.4761 | Yes | ||
| 47 | SNRPB2 | 1870 | 2.965 | 0.4800 | Yes | ||
| 48 | LSM6 | 2345 | 2.621 | 0.4636 | No | ||
| 49 | NOL5A | 2528 | 2.524 | 0.4604 | No | ||
| 50 | NSUN2 | 2547 | 2.512 | 0.4647 | No | ||
| 51 | RBPMS | 2737 | 2.420 | 0.4609 | No | ||
| 52 | EXOSC3 | 2851 | 2.367 | 0.4606 | No | ||
| 53 | SF3A2 | 2878 | 2.355 | 0.4642 | No | ||
| 54 | IGF2BP3 | 2932 | 2.328 | 0.4665 | No | ||
| 55 | DDX54 | 3162 | 2.236 | 0.4605 | No | ||
| 56 | KHDRBS1 | 3244 | 2.202 | 0.4613 | No | ||
| 57 | SETX | 3349 | 2.161 | 0.4609 | No | ||
| 58 | HNRPD | 3388 | 2.146 | 0.4636 | No | ||
| 59 | RPS14 | 3760 | 2.005 | 0.4507 | No | ||
| 60 | DHX38 | 3868 | 1.969 | 0.4498 | No | ||
| 61 | GEMIN6 | 4147 | 1.881 | 0.4408 | No | ||
| 62 | METTL1 | 4334 | 1.816 | 0.4360 | No | ||
| 63 | CPSF3 | 4731 | 1.689 | 0.4213 | No | ||
| 64 | SNW1 | 4819 | 1.662 | 0.4207 | No | ||
| 65 | HNRPF | 5130 | 1.564 | 0.4097 | No | ||
| 66 | SNRPD3 | 5251 | 1.529 | 0.4073 | No | ||
| 67 | ASCC3L1 | 5367 | 1.497 | 0.4051 | No | ||
| 68 | CDC2L1 | 5392 | 1.492 | 0.4070 | No | ||
| 69 | PHF5A | 5598 | 1.435 | 0.4005 | No | ||
| 70 | NOLA2 | 5953 | 1.344 | 0.3870 | No | ||
| 71 | EFTUD2 | 6040 | 1.326 | 0.3858 | No | ||
| 72 | GEMIN7 | 6429 | 1.233 | 0.3705 | No | ||
| 73 | SFRS9 | 6490 | 1.219 | 0.3702 | No | ||
| 74 | AGGF1 | 6809 | 1.146 | 0.3580 | No | ||
| 75 | U2AF1 | 6951 | 1.111 | 0.3538 | No | ||
| 76 | GEMIN5 | 7590 | 0.965 | 0.3265 | No | ||
| 77 | CSTF3 | 9224 | 0.588 | 0.2528 | No | ||
| 78 | PPAN | 9522 | 0.516 | 0.2402 | No | ||
| 79 | TXNL4A | 9551 | 0.507 | 0.2400 | No | ||
| 80 | PRPF18 | 9795 | 0.439 | 0.2297 | No | ||
| 81 | LSM5 | 9941 | 0.398 | 0.2239 | No | ||
| 82 | SNRPD1 | 10547 | 0.229 | 0.1966 | No | ||
| 83 | SARS | 10642 | 0.203 | 0.1927 | No | ||
| 84 | RBMY1A1 | 10801 | 0.151 | 0.1858 | No | ||
| 85 | UPF1 | 12211 | -0.278 | 0.1217 | No | ||
| 86 | USP39 | 12617 | -0.398 | 0.1040 | No | ||
| 87 | HNRPUL1 | 12768 | -0.442 | 0.0980 | No | ||
| 88 | SNRPF | 13159 | -0.548 | 0.0812 | No | ||
| 89 | CPSF1 | 13281 | -0.581 | 0.0768 | No | ||
| 90 | LSM4 | 13433 | -0.630 | 0.0712 | No | ||
| 91 | WDR57 | 13709 | -0.703 | 0.0600 | No | ||
| 92 | LSM3 | 13827 | -0.731 | 0.0561 | No | ||
| 93 | SNRPN | 13906 | -0.754 | 0.0541 | No | ||
| 94 | PRPF8 | 13923 | -0.758 | 0.0549 | No | ||
| 95 | DHX16 | 14862 | -1.004 | 0.0139 | No | ||
| 96 | ADAT1 | 14886 | -1.011 | 0.0149 | No | ||
| 97 | EXOSC2 | 15127 | -1.081 | 0.0061 | No | ||
| 98 | INTS6 | 15429 | -1.163 | -0.0053 | No | ||
| 99 | GEMIN4 | 15491 | -1.180 | -0.0057 | No | ||
| 100 | DBR1 | 15687 | -1.230 | -0.0121 | No | ||
| 101 | AARS | 16038 | -1.319 | -0.0255 | No | ||
| 102 | SRPK1 | 16374 | -1.408 | -0.0380 | No | ||
| 103 | SIP1 | 16481 | -1.436 | -0.0399 | No | ||
| 104 | INTS5 | 16628 | -1.475 | -0.0436 | No | ||
| 105 | DHX8 | 17049 | -1.600 | -0.0596 | No | ||
| 106 | INTS10 | 17377 | -1.713 | -0.0711 | No | ||
| 107 | BICD1 | 17521 | -1.757 | -0.0741 | No | ||
| 108 | RBMS1 | 17540 | -1.765 | -0.0713 | No | ||
| 109 | RBM3 | 18079 | -1.950 | -0.0920 | No | ||
| 110 | PPARGC1A | 18084 | -1.952 | -0.0882 | No | ||
| 111 | SF3A1 | 18423 | -2.087 | -0.0995 | No | ||
| 112 | DDX17 | 18811 | -2.253 | -0.1126 | No | ||
| 113 | DDX39 | 18913 | -2.299 | -0.1126 | No | ||
| 114 | EXOSC7 | 19181 | -2.434 | -0.1198 | No | ||
| 115 | FBL | 19202 | -2.447 | -0.1158 | No | ||
| 116 | FARS2 | 19263 | -2.480 | -0.1135 | No | ||
| 117 | SF1 | 19340 | -2.515 | -0.1118 | No | ||
| 118 | SFRS4 | 19374 | -2.533 | -0.1082 | No | ||
| 119 | ZNF346 | 19604 | -2.662 | -0.1133 | No | ||
| 120 | RNPS1 | 19627 | -2.669 | -0.1088 | No | ||
| 121 | BCAS2 | 19653 | -2.682 | -0.1045 | No | ||
| 122 | DDX23 | 19698 | -2.710 | -0.1010 | No | ||
| 123 | LSM1 | 19922 | -2.842 | -0.1054 | No | ||
| 124 | PTBP1 | 19927 | -2.846 | -0.0998 | No | ||
| 125 | CPSF3L | 20051 | -2.927 | -0.0995 | No | ||
| 126 | RBMS2 | 20172 | -3.013 | -0.0988 | No | ||
| 127 | SNRP70 | 20333 | -3.122 | -0.0998 | No | ||
| 128 | ZNF638 | 20334 | -3.123 | -0.0935 | No | ||
| 129 | SLBP | 20555 | -3.309 | -0.0968 | No | ||
| 130 | PRPF31 | 20568 | -3.325 | -0.0906 | No | ||
| 131 | SF3B3 | 20619 | -3.372 | -0.0860 | No | ||
| 132 | SNRPB | 20743 | -3.502 | -0.0845 | No | ||
| 133 | SNRPD2 | 20816 | -3.588 | -0.0805 | No | ||
| 134 | SF3B2 | 20976 | -3.783 | -0.0801 | No | ||
| 135 | SPOP | 21064 | -3.916 | -0.0761 | No | ||
| 136 | SRPK2 | 21253 | -4.263 | -0.0760 | No | ||
| 137 | CDC40 | 21294 | -4.345 | -0.0690 | No | ||
| 138 | RBM5 | 21505 | -4.898 | -0.0686 | No | ||
| 139 | SFRS8 | 21582 | -5.193 | -0.0615 | No | ||
| 140 | NONO | 21732 | -5.944 | -0.0562 | No | ||
| 141 | CSTF1 | 21753 | -6.103 | -0.0447 | No | ||
| 142 | KHSRP | 21778 | -6.332 | -0.0329 | No | ||
| 143 | INTS3 | 21876 | -8.418 | -0.0202 | No | ||
| 144 | INTS7 | 21921 | -11.484 | 0.0012 | No |
