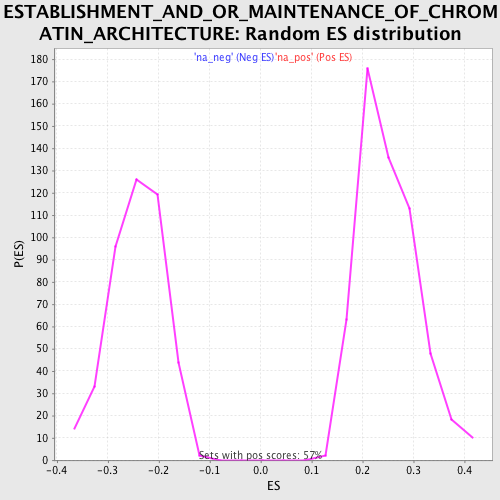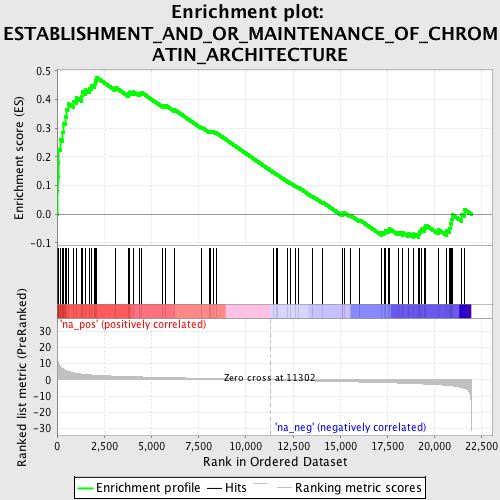
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set01_ATM_minus_versus_SCID |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_pos |
| GeneSet | ESTABLISHMENT_AND_OR_MAINTENANCE_OF_CHROMATIN_ARCHITECTURE |
| Enrichment Score (ES) | 0.47614762 |
| Normalized Enrichment Score (NES) | 1.9037712 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.048917703 |
| FWER p-Value | 0.583 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | SMARCE1 | 7 | 16.881 | 0.0804 | Yes | ||
| 2 | ASF1A | 46 | 11.089 | 0.1316 | Yes | ||
| 3 | EZH2 | 58 | 10.502 | 0.1813 | Yes | ||
| 4 | SET | 97 | 9.283 | 0.2239 | Yes | ||
| 5 | NASP | 168 | 8.269 | 0.2602 | Yes | ||
| 6 | SMARCC1 | 305 | 6.770 | 0.2864 | Yes | ||
| 7 | NAP1L1 | 321 | 6.673 | 0.3176 | Yes | ||
| 8 | HUWE1 | 463 | 5.818 | 0.3389 | Yes | ||
| 9 | WHSC1L1 | 477 | 5.742 | 0.3658 | Yes | ||
| 10 | HDAC5 | 577 | 5.215 | 0.3862 | Yes | ||
| 11 | TLK2 | 861 | 4.415 | 0.3943 | Yes | ||
| 12 | RPS6KA5 | 1025 | 4.034 | 0.4062 | Yes | ||
| 13 | SIRT1 | 1309 | 3.548 | 0.4102 | Yes | ||
| 14 | ACTL6A | 1321 | 3.529 | 0.4265 | Yes | ||
| 15 | HMGB1 | 1523 | 3.287 | 0.4331 | Yes | ||
| 16 | HMGA1 | 1732 | 3.086 | 0.4383 | Yes | ||
| 17 | HIRIP3 | 1842 | 2.988 | 0.4476 | Yes | ||
| 18 | TSSK6 | 2004 | 2.845 | 0.4538 | Yes | ||
| 19 | SYCP3 | 2020 | 2.835 | 0.4667 | Yes | ||
| 20 | SUPT16H | 2103 | 2.765 | 0.4761 | Yes | ||
| 21 | HDAC4 | 3069 | 2.271 | 0.4429 | No | ||
| 22 | HDAC2 | 3774 | 2.000 | 0.4203 | No | ||
| 23 | GCN5L2 | 3858 | 1.973 | 0.4259 | No | ||
| 24 | MAP3K12 | 4056 | 1.908 | 0.4260 | No | ||
| 25 | PRMT7 | 4350 | 1.812 | 0.4213 | No | ||
| 26 | CHAF1B | 4460 | 1.779 | 0.4248 | No | ||
| 27 | SUV39H2 | 5592 | 1.435 | 0.3800 | No | ||
| 28 | PRMT8 | 5751 | 1.396 | 0.3794 | No | ||
| 29 | UBE2N | 6220 | 1.280 | 0.3641 | No | ||
| 30 | H1FNT | 7665 | 0.947 | 0.3027 | No | ||
| 31 | HDAC1 | 8081 | 0.851 | 0.2878 | No | ||
| 32 | RSF1 | 8138 | 0.837 | 0.2892 | No | ||
| 33 | TNP1 | 8262 | 0.810 | 0.2874 | No | ||
| 34 | SMARCD1 | 8437 | 0.772 | 0.2832 | No | ||
| 35 | NAP1L3 | 11477 | -0.060 | 0.1446 | No | ||
| 36 | SIRT2 | 11642 | -0.107 | 0.1376 | No | ||
| 37 | BNIP3 | 11691 | -0.124 | 0.1360 | No | ||
| 38 | JMJD2A | 12228 | -0.281 | 0.1128 | No | ||
| 39 | NAP1L2 | 12379 | -0.325 | 0.1075 | No | ||
| 40 | HDAC11 | 12639 | -0.405 | 0.0976 | No | ||
| 41 | EHMT1 | 12815 | -0.453 | 0.0918 | No | ||
| 42 | MTA2 | 13520 | -0.656 | 0.0627 | No | ||
| 43 | ARID1A | 14066 | -0.796 | 0.0416 | No | ||
| 44 | HDAC7A | 15114 | -1.077 | -0.0011 | No | ||
| 45 | SIRT4 | 15115 | -1.078 | 0.0041 | No | ||
| 46 | HDAC8 | 15218 | -1.106 | 0.0047 | No | ||
| 47 | NAP1L4 | 15522 | -1.189 | -0.0035 | No | ||
| 48 | CARM1 | 16044 | -1.321 | -0.0210 | No | ||
| 49 | INOC1 | 17185 | -1.643 | -0.0652 | No | ||
| 50 | MYST1 | 17322 | -1.690 | -0.0634 | No | ||
| 51 | MYST3 | 17384 | -1.715 | -0.0580 | No | ||
| 52 | SAFB | 17541 | -1.765 | -0.0567 | No | ||
| 53 | TAF6L | 17614 | -1.789 | -0.0514 | No | ||
| 54 | PPARGC1A | 18084 | -1.952 | -0.0635 | No | ||
| 55 | SIRT5 | 18305 | -2.041 | -0.0638 | No | ||
| 56 | HTATIP | 18602 | -2.156 | -0.0670 | No | ||
| 57 | HMGA2 | 18892 | -2.290 | -0.0693 | No | ||
| 58 | CREBBP | 19159 | -2.422 | -0.0699 | No | ||
| 59 | TLK1 | 19196 | -2.443 | -0.0599 | No | ||
| 60 | HELLS | 19283 | -2.494 | -0.0519 | No | ||
| 61 | SMARCA5 | 19457 | -2.580 | -0.0475 | No | ||
| 62 | NSD1 | 19541 | -2.628 | -0.0387 | No | ||
| 63 | MYST4 | 20200 | -3.031 | -0.0543 | No | ||
| 64 | HDAC3 | 20635 | -3.386 | -0.0579 | No | ||
| 65 | HDAC10 | 20807 | -3.579 | -0.0487 | No | ||
| 66 | SATB1 | 20856 | -3.642 | -0.0334 | No | ||
| 67 | RBM14 | 20888 | -3.671 | -0.0173 | No | ||
| 68 | RBBP4 | 20957 | -3.754 | -0.0025 | No | ||
| 69 | SMARCC2 | 21446 | -4.732 | -0.0022 | No | ||
| 70 | PHB | 21598 | -5.236 | 0.0160 | No |