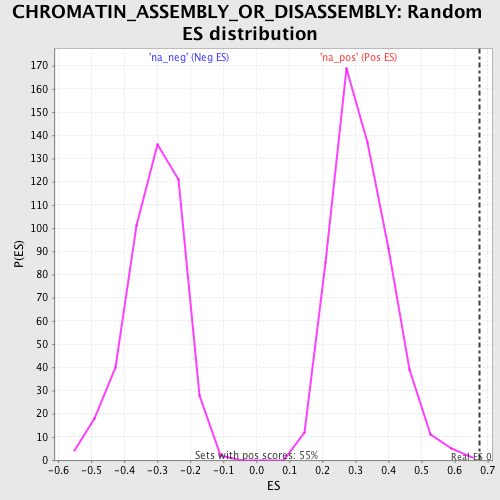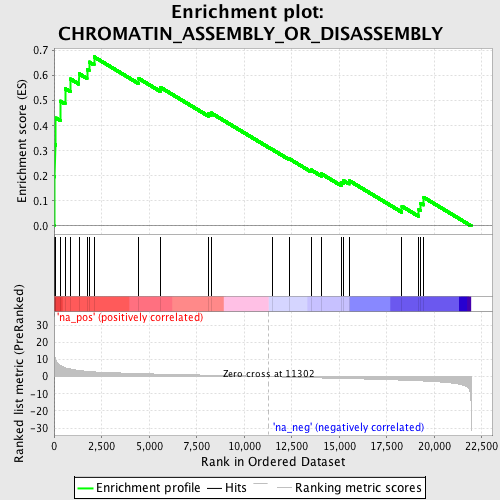
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set01_ATM_minus_versus_SCID |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_pos |
| GeneSet | CHROMATIN_ASSEMBLY_OR_DISASSEMBLY |
| Enrichment Score (ES) | 0.67510283 |
| Normalized Enrichment Score (NES) | 2.1172996 |
| Nominal p-value | 0.0018181818 |
| FDR q-value | 0.004071269 |
| FWER p-Value | 0.029 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | SMARCE1 | 7 | 16.881 | 0.1970 | Yes | ||
| 2 | ASF1A | 46 | 11.089 | 0.3248 | Yes | ||
| 3 | SET | 97 | 9.283 | 0.4310 | Yes | ||
| 4 | NAP1L1 | 321 | 6.673 | 0.4988 | Yes | ||
| 5 | HDAC5 | 577 | 5.215 | 0.5481 | Yes | ||
| 6 | TLK2 | 861 | 4.415 | 0.5868 | Yes | ||
| 7 | SIRT1 | 1309 | 3.548 | 0.6079 | Yes | ||
| 8 | HMGA1 | 1732 | 3.086 | 0.6247 | Yes | ||
| 9 | HIRIP3 | 1842 | 2.988 | 0.6546 | Yes | ||
| 10 | SUPT16H | 2103 | 2.765 | 0.6751 | Yes | ||
| 11 | CHAF1B | 4460 | 1.779 | 0.5884 | No | ||
| 12 | SUV39H2 | 5592 | 1.435 | 0.5536 | No | ||
| 13 | RSF1 | 8138 | 0.837 | 0.4473 | No | ||
| 14 | TNP1 | 8262 | 0.810 | 0.4512 | No | ||
| 15 | NAP1L3 | 11477 | -0.060 | 0.3053 | No | ||
| 16 | NAP1L2 | 12379 | -0.325 | 0.2680 | No | ||
| 17 | MTA2 | 13520 | -0.656 | 0.2236 | No | ||
| 18 | ARID1A | 14066 | -0.796 | 0.2081 | No | ||
| 19 | SIRT4 | 15115 | -1.078 | 0.1729 | No | ||
| 20 | HDAC8 | 15218 | -1.106 | 0.1811 | No | ||
| 21 | NAP1L4 | 15522 | -1.189 | 0.1812 | No | ||
| 22 | SIRT5 | 18305 | -2.041 | 0.0781 | No | ||
| 23 | TLK1 | 19196 | -2.443 | 0.0661 | No | ||
| 24 | HELLS | 19283 | -2.494 | 0.0913 | No | ||
| 25 | SMARCA5 | 19457 | -2.580 | 0.1136 | No |