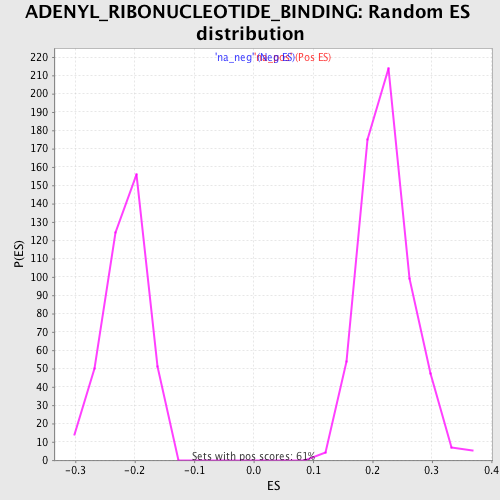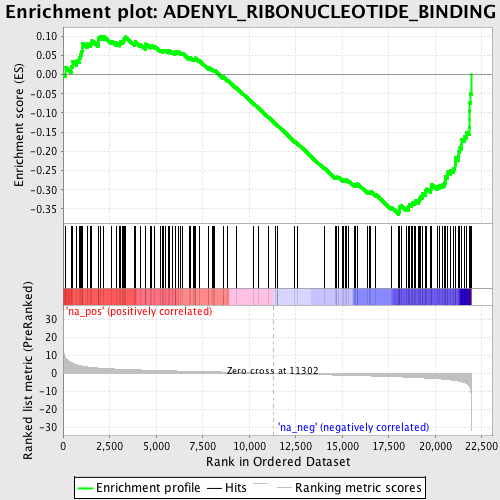
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set01_ATM_minus_versus_SCID |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_neg |
| GeneSet | ADENYL_RIBONUCLEOTIDE_BINDING |
| Enrichment Score (ES) | -0.3644462 |
| Normalized Enrichment Score (NES) | -1.6924162 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.33654875 |
| FWER p-Value | 0.998 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | ABCB7 | 130 | 8.716 | 0.0188 | No | ||
| 2 | RFC1 | 445 | 5.920 | 0.0212 | No | ||
| 3 | ABCF1 | 522 | 5.498 | 0.0334 | No | ||
| 4 | SMC4 | 744 | 4.673 | 0.0365 | No | ||
| 5 | TLK2 | 861 | 4.415 | 0.0437 | No | ||
| 6 | N4BP2 | 911 | 4.272 | 0.0536 | No | ||
| 7 | HSP90AA1 | 1006 | 4.060 | 0.0608 | No | ||
| 8 | RPS6KA5 | 1025 | 4.034 | 0.0715 | No | ||
| 9 | WNK1 | 1044 | 3.998 | 0.0820 | No | ||
| 10 | TRNT1 | 1312 | 3.537 | 0.0798 | No | ||
| 11 | DYRK2 | 1469 | 3.348 | 0.0822 | No | ||
| 12 | NLK | 1550 | 3.262 | 0.0878 | No | ||
| 13 | ABCC1 | 1878 | 2.960 | 0.0812 | No | ||
| 14 | NVL | 1897 | 2.940 | 0.0887 | No | ||
| 15 | LATS2 | 1924 | 2.919 | 0.0958 | No | ||
| 16 | TSSK6 | 2004 | 2.845 | 0.1003 | No | ||
| 17 | DYRK3 | 2182 | 2.713 | 0.0998 | No | ||
| 18 | ABCB8 | 2621 | 2.468 | 0.0868 | No | ||
| 19 | TSSK4 | 2860 | 2.363 | 0.0826 | No | ||
| 20 | PINK1 | 3037 | 2.282 | 0.0810 | No | ||
| 21 | MAP3K6 | 3076 | 2.268 | 0.0857 | No | ||
| 22 | GABARAPL2 | 3198 | 2.220 | 0.0864 | No | ||
| 23 | TSSK3 | 3248 | 2.201 | 0.0904 | No | ||
| 24 | MYO3A | 3302 | 2.181 | 0.0942 | No | ||
| 25 | GCK | 3354 | 2.159 | 0.0980 | No | ||
| 26 | PEX6 | 3839 | 1.978 | 0.0814 | No | ||
| 27 | DHX38 | 3868 | 1.969 | 0.0857 | No | ||
| 28 | ICK | 4152 | 1.880 | 0.0781 | No | ||
| 29 | LATS1 | 4409 | 1.798 | 0.0715 | No | ||
| 30 | ACVR1 | 4432 | 1.789 | 0.0755 | No | ||
| 31 | MSH2 | 4451 | 1.783 | 0.0798 | No | ||
| 32 | NEK6 | 4682 | 1.707 | 0.0741 | No | ||
| 33 | EIF2B2 | 4732 | 1.688 | 0.0766 | No | ||
| 34 | IRAK3 | 4901 | 1.637 | 0.0736 | No | ||
| 35 | ABCB6 | 5218 | 1.538 | 0.0635 | No | ||
| 36 | DMC1 | 5355 | 1.502 | 0.0615 | No | ||
| 37 | CDC2L1 | 5392 | 1.492 | 0.0641 | No | ||
| 38 | HSPA4L | 5498 | 1.459 | 0.0634 | No | ||
| 39 | TRIB1 | 5655 | 1.422 | 0.0603 | No | ||
| 40 | TNK1 | 5689 | 1.411 | 0.0628 | No | ||
| 41 | BTK | 5881 | 1.361 | 0.0579 | No | ||
| 42 | DDX25 | 6012 | 1.331 | 0.0557 | No | ||
| 43 | BCKDK | 6019 | 1.330 | 0.0592 | No | ||
| 44 | MYO6 | 6056 | 1.322 | 0.0613 | No | ||
| 45 | NEK11 | 6176 | 1.290 | 0.0595 | No | ||
| 46 | MAST1 | 6331 | 1.254 | 0.0560 | No | ||
| 47 | MKNK1 | 6427 | 1.234 | 0.0552 | No | ||
| 48 | WNK4 | 6772 | 1.154 | 0.0427 | No | ||
| 49 | PIM1 | 6823 | 1.141 | 0.0436 | No | ||
| 50 | ABCF2 | 6989 | 1.103 | 0.0392 | No | ||
| 51 | BMPR1B | 7071 | 1.082 | 0.0386 | No | ||
| 52 | WNK2 | 7088 | 1.076 | 0.0409 | No | ||
| 53 | PRKCI | 7102 | 1.072 | 0.0433 | No | ||
| 54 | ABCD4 | 7323 | 1.023 | 0.0361 | No | ||
| 55 | CDC42BPB | 7809 | 0.916 | 0.0165 | No | ||
| 56 | ABCC6 | 7832 | 0.911 | 0.0181 | No | ||
| 57 | APRT | 8008 | 0.868 | 0.0125 | No | ||
| 58 | PEX1 | 8089 | 0.849 | 0.0113 | No | ||
| 59 | PGK2 | 8156 | 0.833 | 0.0106 | No | ||
| 60 | DGKE | 8593 | 0.736 | -0.0073 | No | ||
| 61 | ABCG2 | 8597 | 0.735 | -0.0053 | No | ||
| 62 | ABCC8 | 8854 | 0.673 | -0.0152 | No | ||
| 63 | RAD51 | 9305 | 0.569 | -0.0342 | No | ||
| 64 | MSH6 | 10207 | 0.325 | -0.0746 | No | ||
| 65 | WNK3 | 10497 | 0.241 | -0.0871 | No | ||
| 66 | MOV10L1 | 11036 | 0.078 | -0.1116 | No | ||
| 67 | SPHK1 | 11395 | -0.030 | -0.1279 | No | ||
| 68 | ERN2 | 11492 | -0.065 | -0.1321 | No | ||
| 69 | OXSR1 | 11509 | -0.069 | -0.1327 | No | ||
| 70 | CFTR | 11521 | -0.072 | -0.1330 | No | ||
| 71 | TSSK2 | 12458 | -0.351 | -0.1749 | No | ||
| 72 | CDC42BPG | 12609 | -0.396 | -0.1807 | No | ||
| 73 | ATP7B | 14033 | -0.788 | -0.2437 | No | ||
| 74 | ABCG1 | 14629 | -0.938 | -0.2683 | No | ||
| 75 | NME1 | 14665 | -0.949 | -0.2672 | No | ||
| 76 | GCLC | 14715 | -0.961 | -0.2667 | No | ||
| 77 | STK38L | 14783 | -0.981 | -0.2670 | No | ||
| 78 | ACVRL1 | 15021 | -1.052 | -0.2749 | No | ||
| 79 | STK4 | 15042 | -1.057 | -0.2728 | No | ||
| 80 | MKNK2 | 15165 | -1.091 | -0.2753 | No | ||
| 81 | ABCA4 | 15204 | -1.102 | -0.2739 | No | ||
| 82 | GSG2 | 15359 | -1.142 | -0.2777 | No | ||
| 83 | STK36 | 15681 | -1.229 | -0.2890 | No | ||
| 84 | MAP4K3 | 15693 | -1.231 | -0.2860 | No | ||
| 85 | NUAK2 | 15806 | -1.262 | -0.2875 | No | ||
| 86 | SEPHS1 | 15828 | -1.268 | -0.2849 | No | ||
| 87 | SRPK1 | 16374 | -1.408 | -0.3059 | No | ||
| 88 | SNF1LK | 16473 | -1.434 | -0.3063 | No | ||
| 89 | ACVR1B | 16502 | -1.442 | -0.3035 | No | ||
| 90 | CDC42BPA | 16810 | -1.528 | -0.3132 | No | ||
| 91 | VPS4A | 17666 | -1.804 | -0.3473 | No | ||
| 92 | TPX2 | 18041 | -1.938 | -0.3589 | Yes | ||
| 93 | GALK1 | 18053 | -1.942 | -0.3539 | Yes | ||
| 94 | RFC2 | 18059 | -1.944 | -0.3486 | Yes | ||
| 95 | BRSK2 | 18082 | -1.952 | -0.3441 | Yes | ||
| 96 | ABCC5 | 18155 | -1.979 | -0.3418 | Yes | ||
| 97 | DMPK | 18432 | -2.092 | -0.3485 | Yes | ||
| 98 | ABCA7 | 18553 | -2.140 | -0.3479 | Yes | ||
| 99 | ACTA1 | 18569 | -2.146 | -0.3425 | Yes | ||
| 100 | ABCB11 | 18587 | -2.150 | -0.3372 | Yes | ||
| 101 | MARK1 | 18743 | -2.222 | -0.3380 | Yes | ||
| 102 | DAPK3 | 18783 | -2.238 | -0.3334 | Yes | ||
| 103 | TTN | 18906 | -2.297 | -0.3325 | Yes | ||
| 104 | MYH10 | 18956 | -2.317 | -0.3281 | Yes | ||
| 105 | ATP7A | 19110 | -2.393 | -0.3284 | Yes | ||
| 106 | MAP4K5 | 19152 | -2.418 | -0.3234 | Yes | ||
| 107 | TLK1 | 19196 | -2.443 | -0.3184 | Yes | ||
| 108 | STK11 | 19290 | -2.495 | -0.3156 | Yes | ||
| 109 | ERCC6 | 19324 | -2.511 | -0.3100 | Yes | ||
| 110 | SMARCA5 | 19457 | -2.580 | -0.3087 | Yes | ||
| 111 | ERN1 | 19490 | -2.602 | -0.3028 | Yes | ||
| 112 | ABCD2 | 19551 | -2.634 | -0.2980 | Yes | ||
| 113 | MAP4K2 | 19737 | -2.734 | -0.2987 | Yes | ||
| 114 | BMPR1A | 19772 | -2.752 | -0.2925 | Yes | ||
| 115 | ABCA3 | 19795 | -2.760 | -0.2857 | Yes | ||
| 116 | DGKZ | 20098 | -2.964 | -0.2911 | Yes | ||
| 117 | DAPK2 | 20215 | -3.039 | -0.2878 | Yes | ||
| 118 | STK17B | 20367 | -3.150 | -0.2857 | Yes | ||
| 119 | SNF1LK2 | 20484 | -3.260 | -0.2818 | Yes | ||
| 120 | RPS6KA4 | 20524 | -3.290 | -0.2742 | Yes | ||
| 121 | TOR1A | 20535 | -3.297 | -0.2653 | Yes | ||
| 122 | MAST2 | 20654 | -3.411 | -0.2611 | Yes | ||
| 123 | ABCA2 | 20679 | -3.426 | -0.2524 | Yes | ||
| 124 | MARK2 | 20828 | -3.598 | -0.2490 | Yes | ||
| 125 | STK38 | 20973 | -3.779 | -0.2449 | Yes | ||
| 126 | SARS2 | 21057 | -3.905 | -0.2376 | Yes | ||
| 127 | TGFBR1 | 21067 | -3.917 | -0.2269 | Yes | ||
| 128 | SNRK | 21101 | -3.967 | -0.2171 | Yes | ||
| 129 | SRPK2 | 21253 | -4.263 | -0.2119 | Yes | ||
| 130 | ABCC3 | 21266 | -4.295 | -0.2003 | Yes | ||
| 131 | STK3 | 21322 | -4.396 | -0.1903 | Yes | ||
| 132 | PPP2R4 | 21407 | -4.609 | -0.1810 | Yes | ||
| 133 | PRKACB | 21416 | -4.640 | -0.1682 | Yes | ||
| 134 | MINK1 | 21558 | -5.091 | -0.1602 | Yes | ||
| 135 | TP53 | 21670 | -5.532 | -0.1496 | Yes | ||
| 136 | ABL1 | 21836 | -7.213 | -0.1367 | Yes | ||
| 137 | MYH9 | 21845 | -7.447 | -0.1159 | Yes | ||
| 138 | MYO9B | 21847 | -7.464 | -0.0947 | Yes | ||
| 139 | MAP4K4 | 21855 | -7.660 | -0.0733 | Yes | ||
| 140 | PXK | 21882 | -8.649 | -0.0499 | Yes | ||
| 141 | DAPK1 | 21946 | -18.608 | 0.0000 | Yes |