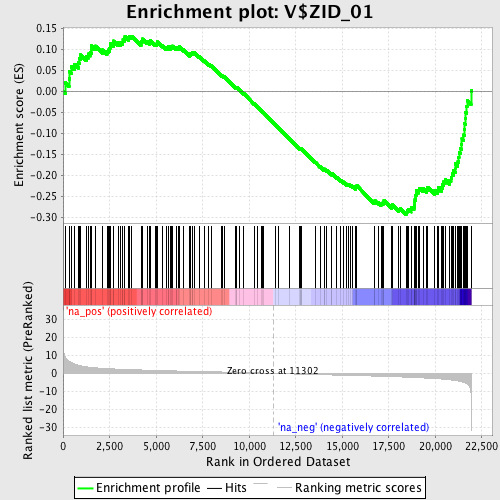
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set01_ATM_minus_versus_SCID |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_neg |
| GeneSet | V$ZID_01 |
| Enrichment Score (ES) | -0.2933322 |
| Normalized Enrichment Score (NES) | -1.3754424 |
| Nominal p-value | 0.02173913 |
| FDR q-value | 0.36106423 |
| FWER p-Value | 0.999 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | KPNA4 | 112 | 9.058 | 0.0203 | No | ||
| 2 | DDX5 | 320 | 6.691 | 0.0296 | No | ||
| 3 | COMMD10 | 345 | 6.508 | 0.0468 | No | ||
| 4 | MBNL1 | 447 | 5.895 | 0.0587 | No | ||
| 5 | ADSS | 623 | 5.028 | 0.0648 | No | ||
| 6 | SKIL | 836 | 4.455 | 0.0676 | No | ||
| 7 | TCF4 | 887 | 4.351 | 0.0775 | No | ||
| 8 | ATP2A2 | 947 | 4.197 | 0.0866 | No | ||
| 9 | RBPSUH | 1244 | 3.647 | 0.0833 | No | ||
| 10 | LRCH4 | 1340 | 3.510 | 0.0888 | No | ||
| 11 | SEMA6C | 1448 | 3.373 | 0.0933 | No | ||
| 12 | HMGB1 | 1523 | 3.287 | 0.0992 | No | ||
| 13 | SLC35A1 | 1531 | 3.279 | 0.1081 | No | ||
| 14 | PA2G4 | 1729 | 3.087 | 0.1077 | No | ||
| 15 | SUPT16H | 2103 | 2.765 | 0.0984 | No | ||
| 16 | STXBP6 | 2360 | 2.609 | 0.0939 | No | ||
| 17 | ACTN3 | 2424 | 2.577 | 0.0983 | No | ||
| 18 | HOXA3 | 2496 | 2.540 | 0.1022 | No | ||
| 19 | AKAP1 | 2524 | 2.525 | 0.1080 | No | ||
| 20 | TMEFF1 | 2555 | 2.509 | 0.1137 | No | ||
| 21 | MTF1 | 2711 | 2.428 | 0.1134 | No | ||
| 22 | CHD4 | 2724 | 2.423 | 0.1197 | No | ||
| 23 | TLX3 | 2959 | 2.318 | 0.1154 | No | ||
| 24 | TIAM1 | 3094 | 2.261 | 0.1156 | No | ||
| 25 | GABARAPL2 | 3198 | 2.220 | 0.1172 | No | ||
| 26 | CACNG2 | 3216 | 2.215 | 0.1226 | No | ||
| 27 | PCF11 | 3289 | 2.185 | 0.1254 | No | ||
| 28 | DDIT4L | 3323 | 2.169 | 0.1300 | No | ||
| 29 | PLXNA3 | 3510 | 2.098 | 0.1274 | No | ||
| 30 | PSMD8 | 3556 | 2.078 | 0.1311 | No | ||
| 31 | CHRD | 3680 | 2.033 | 0.1312 | No | ||
| 32 | FKBP14 | 4187 | 1.866 | 0.1132 | No | ||
| 33 | PKIB | 4190 | 1.865 | 0.1184 | No | ||
| 34 | UST | 4243 | 1.850 | 0.1212 | No | ||
| 35 | MARCKS | 4285 | 1.832 | 0.1245 | No | ||
| 36 | FGFR1 | 4520 | 1.764 | 0.1187 | No | ||
| 37 | LBX1 | 4663 | 1.710 | 0.1170 | No | ||
| 38 | ZIC4 | 4675 | 1.708 | 0.1213 | No | ||
| 39 | B4GALT2 | 4968 | 1.618 | 0.1124 | No | ||
| 40 | MPP3 | 5042 | 1.592 | 0.1135 | No | ||
| 41 | HR | 5052 | 1.589 | 0.1176 | No | ||
| 42 | ZBTB33 | 5324 | 1.511 | 0.1094 | No | ||
| 43 | PAX6 | 5532 | 1.451 | 0.1040 | No | ||
| 44 | RAB2 | 5659 | 1.420 | 0.1022 | No | ||
| 45 | MYH6 | 5662 | 1.420 | 0.1061 | No | ||
| 46 | C1QTNF6 | 5794 | 1.383 | 0.1039 | No | ||
| 47 | DPF3 | 5833 | 1.374 | 0.1061 | No | ||
| 48 | BTK | 5881 | 1.361 | 0.1077 | No | ||
| 49 | CAPNS1 | 6077 | 1.317 | 0.1025 | No | ||
| 50 | PLP1 | 6098 | 1.313 | 0.1052 | No | ||
| 51 | UBE2N | 6220 | 1.280 | 0.1033 | No | ||
| 52 | NLGN3 | 6230 | 1.278 | 0.1065 | No | ||
| 53 | ISL1 | 6476 | 1.223 | 0.0987 | No | ||
| 54 | PSD | 6808 | 1.147 | 0.0867 | No | ||
| 55 | LIN28 | 6829 | 1.139 | 0.0890 | No | ||
| 56 | CSPG2 | 6866 | 1.130 | 0.0905 | No | ||
| 57 | HOXB9 | 6942 | 1.112 | 0.0902 | No | ||
| 58 | DMD | 6960 | 1.109 | 0.0925 | No | ||
| 59 | ZIC1 | 7036 | 1.090 | 0.0921 | No | ||
| 60 | PGF | 7302 | 1.028 | 0.0829 | No | ||
| 61 | GGN | 7592 | 0.965 | 0.0723 | No | ||
| 62 | KCNE1L | 7825 | 0.912 | 0.0642 | No | ||
| 63 | CNTN1 | 7947 | 0.884 | 0.0612 | No | ||
| 64 | PLA2G2F | 8534 | 0.752 | 0.0364 | No | ||
| 65 | MRPL40 | 8576 | 0.740 | 0.0366 | No | ||
| 66 | ERBB3 | 8646 | 0.726 | 0.0354 | No | ||
| 67 | SESN3 | 9271 | 0.576 | 0.0084 | No | ||
| 68 | TNKS1BP1 | 9313 | 0.568 | 0.0081 | No | ||
| 69 | PPP2R2B | 9329 | 0.564 | 0.0090 | No | ||
| 70 | GRIK3 | 9485 | 0.527 | 0.0034 | No | ||
| 71 | THPO | 9716 | 0.460 | -0.0059 | No | ||
| 72 | HTR1B | 10261 | 0.312 | -0.0299 | No | ||
| 73 | CMYA1 | 10445 | 0.258 | -0.0376 | No | ||
| 74 | PIPOX | 10685 | 0.190 | -0.0481 | No | ||
| 75 | SLC17A3 | 10734 | 0.178 | -0.0498 | No | ||
| 76 | PHOSPHO1 | 10767 | 0.163 | -0.0508 | No | ||
| 77 | TLX1 | 11437 | -0.046 | -0.0813 | No | ||
| 78 | PRELP | 11570 | -0.090 | -0.0871 | No | ||
| 79 | MEOX1 | 12159 | -0.262 | -0.1134 | No | ||
| 80 | ENPP1 | 12681 | -0.416 | -0.1361 | No | ||
| 81 | FBXO24 | 12742 | -0.434 | -0.1377 | No | ||
| 82 | HNRPUL1 | 12768 | -0.442 | -0.1376 | No | ||
| 83 | FAU | 12788 | -0.448 | -0.1372 | No | ||
| 84 | SETBP1 | 12826 | -0.456 | -0.1376 | No | ||
| 85 | TRIM46 | 13543 | -0.661 | -0.1686 | No | ||
| 86 | HIST1H3D | 13822 | -0.729 | -0.1793 | No | ||
| 87 | SPAG8 | 14041 | -0.790 | -0.1871 | No | ||
| 88 | SP4 | 14045 | -0.791 | -0.1850 | No | ||
| 89 | ADAM12 | 14172 | -0.823 | -0.1885 | No | ||
| 90 | CTCF | 14413 | -0.882 | -0.1970 | No | ||
| 91 | NGFR | 14422 | -0.884 | -0.1949 | No | ||
| 92 | MRPL24 | 14711 | -0.961 | -0.2054 | No | ||
| 93 | EVA1 | 14914 | -1.024 | -0.2118 | No | ||
| 94 | ERF | 15068 | -1.064 | -0.2158 | No | ||
| 95 | MECP2 | 15244 | -1.114 | -0.2207 | No | ||
| 96 | DDR2 | 15348 | -1.139 | -0.2223 | No | ||
| 97 | PHTF1 | 15459 | -1.172 | -0.2240 | No | ||
| 98 | ESRRG | 15568 | -1.198 | -0.2256 | No | ||
| 99 | WNT6 | 15730 | -1.240 | -0.2295 | No | ||
| 100 | SCRT2 | 15744 | -1.242 | -0.2266 | No | ||
| 101 | HOXB8 | 15788 | -1.257 | -0.2251 | No | ||
| 102 | RB1CC1 | 16718 | -1.502 | -0.2635 | No | ||
| 103 | DNAJB5 | 16756 | -1.514 | -0.2609 | No | ||
| 104 | LTBP1 | 16926 | -1.562 | -0.2643 | No | ||
| 105 | PAPPA | 17083 | -1.611 | -0.2669 | No | ||
| 106 | BCS1L | 17163 | -1.635 | -0.2660 | No | ||
| 107 | TRAPPC3 | 17220 | -1.657 | -0.2639 | No | ||
| 108 | SDF2 | 17240 | -1.661 | -0.2601 | No | ||
| 109 | KRTCAP2 | 17664 | -1.804 | -0.2744 | No | ||
| 110 | DCAMKL1 | 17672 | -1.809 | -0.2697 | No | ||
| 111 | NELL2 | 18047 | -1.941 | -0.2814 | No | ||
| 112 | SART3 | 18119 | -1.965 | -0.2791 | No | ||
| 113 | CDC25A | 18430 | -2.090 | -0.2875 | Yes | ||
| 114 | DACH1 | 18481 | -2.111 | -0.2838 | Yes | ||
| 115 | RIN2 | 18547 | -2.138 | -0.2808 | Yes | ||
| 116 | ABTB2 | 18693 | -2.203 | -0.2813 | Yes | ||
| 117 | MAB21L1 | 18721 | -2.213 | -0.2763 | Yes | ||
| 118 | AMOTL1 | 18856 | -2.271 | -0.2761 | Yes | ||
| 119 | GCNT3 | 18883 | -2.286 | -0.2708 | Yes | ||
| 120 | CDK5R1 | 18897 | -2.293 | -0.2650 | Yes | ||
| 121 | SUPT6H | 18904 | -2.296 | -0.2588 | Yes | ||
| 122 | DRG1 | 18944 | -2.312 | -0.2541 | Yes | ||
| 123 | ELK3 | 18951 | -2.315 | -0.2479 | Yes | ||
| 124 | FASTK | 18971 | -2.324 | -0.2422 | Yes | ||
| 125 | GFI1 | 18975 | -2.325 | -0.2358 | Yes | ||
| 126 | BAHD1 | 19118 | -2.400 | -0.2356 | Yes | ||
| 127 | MAP4K5 | 19152 | -2.418 | -0.2303 | Yes | ||
| 128 | DFNB31 | 19342 | -2.517 | -0.2319 | Yes | ||
| 129 | NT5C1B | 19547 | -2.631 | -0.2339 | Yes | ||
| 130 | NFYA | 19591 | -2.654 | -0.2284 | Yes | ||
| 131 | BCAR3 | 19953 | -2.858 | -0.2369 | Yes | ||
| 132 | CREB3 | 20115 | -2.975 | -0.2360 | Yes | ||
| 133 | PLAG1 | 20154 | -3.001 | -0.2293 | Yes | ||
| 134 | SERPINI1 | 20341 | -3.130 | -0.2290 | Yes | ||
| 135 | APEX2 | 20380 | -3.157 | -0.2219 | Yes | ||
| 136 | PDCD10 | 20445 | -3.216 | -0.2158 | Yes | ||
| 137 | RPS6KA4 | 20524 | -3.290 | -0.2101 | Yes | ||
| 138 | PHF8 | 20770 | -3.528 | -0.2115 | Yes | ||
| 139 | POLG | 20843 | -3.620 | -0.2046 | Yes | ||
| 140 | CHD2 | 20895 | -3.680 | -0.1966 | Yes | ||
| 141 | MSI2 | 20963 | -3.762 | -0.1891 | Yes | ||
| 142 | HIRA | 21077 | -3.929 | -0.1833 | Yes | ||
| 143 | ITPR3 | 21081 | -3.940 | -0.1723 | Yes | ||
| 144 | ODF2 | 21216 | -4.174 | -0.1667 | Yes | ||
| 145 | AP2M1 | 21262 | -4.285 | -0.1568 | Yes | ||
| 146 | TCF8 | 21274 | -4.303 | -0.1452 | Yes | ||
| 147 | FOXP1 | 21341 | -4.453 | -0.1357 | Yes | ||
| 148 | DHX30 | 21418 | -4.645 | -0.1261 | Yes | ||
| 149 | UTX | 21431 | -4.694 | -0.1135 | Yes | ||
| 150 | SNX12 | 21532 | -5.007 | -0.1040 | Yes | ||
| 151 | MYO10 | 21550 | -5.071 | -0.0906 | Yes | ||
| 152 | JMJD1C | 21579 | -5.171 | -0.0773 | Yes | ||
| 153 | MRPL49 | 21618 | -5.304 | -0.0642 | Yes | ||
| 154 | PAFAH1B1 | 21628 | -5.352 | -0.0495 | Yes | ||
| 155 | NEURL | 21692 | -5.636 | -0.0366 | Yes | ||
| 156 | TLN1 | 21707 | -5.706 | -0.0212 | Yes | ||
| 157 | ANXA9 | 21920 | -11.451 | 0.0012 | Yes |