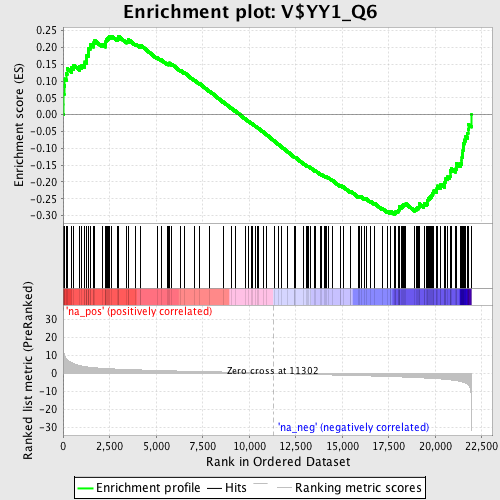
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set01_ATM_minus_versus_SCID |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_neg |
| GeneSet | V$YY1_Q6 |
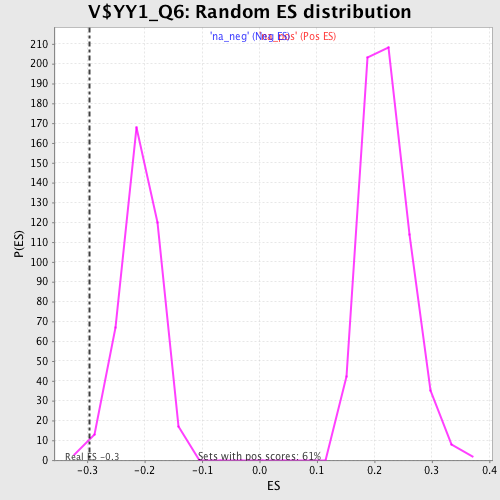
| Enrichment Score (ES) | -0.296565 |
| Normalized Enrichment Score (NES) | -1.4175831 |
| Nominal p-value | 0.0154639175 |
| FDR q-value | 0.4239928 |
| FWER p-Value | 0.989 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | RAB5A | 18 | 13.716 | 0.0311 | No | ||
| 2 | SFRS10 | 19 | 13.401 | 0.0622 | No | ||
| 3 | SNX5 | 51 | 10.860 | 0.0860 | No | ||
| 4 | FKRP | 91 | 9.360 | 0.1060 | No | ||
| 5 | NASP | 168 | 8.269 | 0.1218 | No | ||
| 6 | EIF5 | 212 | 7.562 | 0.1374 | No | ||
| 7 | ELAVL4 | 466 | 5.799 | 0.1392 | No | ||
| 8 | NCOR1 | 566 | 5.268 | 0.1469 | No | ||
| 9 | TCF4 | 887 | 4.351 | 0.1424 | No | ||
| 10 | DEPDC6 | 1007 | 4.058 | 0.1463 | No | ||
| 11 | ARCN1 | 1141 | 3.819 | 0.1491 | No | ||
| 12 | EEF1A1 | 1172 | 3.763 | 0.1565 | No | ||
| 13 | YY1 | 1248 | 3.639 | 0.1615 | No | ||
| 14 | DAP3 | 1259 | 3.622 | 0.1695 | No | ||
| 15 | ZFX | 1275 | 3.602 | 0.1772 | No | ||
| 16 | MORF4L2 | 1347 | 3.499 | 0.1820 | No | ||
| 17 | HIF1A | 1360 | 3.474 | 0.1896 | No | ||
| 18 | ATP5F1 | 1382 | 3.446 | 0.1966 | No | ||
| 19 | POU2F1 | 1447 | 3.374 | 0.2015 | No | ||
| 20 | PTMA | 1471 | 3.346 | 0.2082 | No | ||
| 21 | CIRH1A | 1631 | 3.173 | 0.2083 | No | ||
| 22 | SFPQ | 1655 | 3.149 | 0.2146 | No | ||
| 23 | VAX1 | 1706 | 3.105 | 0.2195 | No | ||
| 24 | RPS8 | 2094 | 2.772 | 0.2082 | No | ||
| 25 | TNKS2 | 2253 | 2.672 | 0.2071 | No | ||
| 26 | UBE3A | 2266 | 2.664 | 0.2128 | No | ||
| 27 | PFN2 | 2299 | 2.641 | 0.2175 | No | ||
| 28 | ATF1 | 2314 | 2.637 | 0.2230 | No | ||
| 29 | MYNN | 2371 | 2.603 | 0.2264 | No | ||
| 30 | TARDBP | 2456 | 2.559 | 0.2285 | No | ||
| 31 | SYT11 | 2490 | 2.544 | 0.2329 | No | ||
| 32 | BTF3 | 2607 | 2.478 | 0.2334 | No | ||
| 33 | BCL11A | 2920 | 2.333 | 0.2245 | No | ||
| 34 | MPDU1 | 2972 | 2.315 | 0.2275 | No | ||
| 35 | TIAL1 | 2987 | 2.307 | 0.2322 | No | ||
| 36 | CACNB2 | 3410 | 2.138 | 0.2178 | No | ||
| 37 | EP300 | 3501 | 2.101 | 0.2186 | No | ||
| 38 | HOXC6 | 3502 | 2.101 | 0.2235 | No | ||
| 39 | DSCAM | 3910 | 1.956 | 0.2093 | No | ||
| 40 | A2BP1 | 4149 | 1.881 | 0.2028 | No | ||
| 41 | USF1 | 4175 | 1.871 | 0.2060 | No | ||
| 42 | HMGB3 | 5053 | 1.589 | 0.1694 | No | ||
| 43 | PPP1R15B | 5272 | 1.524 | 0.1630 | No | ||
| 44 | ZDHHC5 | 5595 | 1.435 | 0.1515 | No | ||
| 45 | PEG3 | 5684 | 1.413 | 0.1508 | No | ||
| 46 | APBB1 | 5695 | 1.410 | 0.1536 | No | ||
| 47 | CCNK | 5837 | 1.373 | 0.1503 | No | ||
| 48 | RAD21 | 6312 | 1.258 | 0.1315 | No | ||
| 49 | MAPT | 6530 | 1.211 | 0.1243 | No | ||
| 50 | WDR13 | 7052 | 1.087 | 0.1029 | No | ||
| 51 | RXRB | 7342 | 1.019 | 0.0920 | No | ||
| 52 | CELSR3 | 7876 | 0.901 | 0.0697 | No | ||
| 53 | PCBP4 | 8607 | 0.733 | 0.0379 | No | ||
| 54 | ATF4 | 9033 | 0.631 | 0.0198 | No | ||
| 55 | MARK3 | 9246 | 0.583 | 0.0115 | No | ||
| 56 | THAP1 | 9786 | 0.442 | -0.0123 | No | ||
| 57 | LYPLA2 | 9957 | 0.395 | -0.0191 | No | ||
| 58 | OSBPL11 | 10124 | 0.347 | -0.0260 | No | ||
| 59 | SNAP25 | 10177 | 0.331 | -0.0276 | No | ||
| 60 | SNURF | 10326 | 0.293 | -0.0337 | No | ||
| 61 | BAMBI | 10442 | 0.259 | -0.0384 | No | ||
| 62 | ABHD1 | 10487 | 0.244 | -0.0398 | No | ||
| 63 | NACA | 10740 | 0.176 | -0.0510 | No | ||
| 64 | UPF3B | 10940 | 0.106 | -0.0599 | No | ||
| 65 | TGM5 | 11343 | -0.013 | -0.0783 | No | ||
| 66 | ZIM2 | 11553 | -0.085 | -0.0877 | No | ||
| 67 | SLC39A7 | 11743 | -0.141 | -0.0960 | No | ||
| 68 | MNT | 12040 | -0.223 | -0.1091 | No | ||
| 69 | MAEA | 12454 | -0.350 | -0.1272 | No | ||
| 70 | WDR34 | 12456 | -0.350 | -0.1265 | No | ||
| 71 | RECK | 12463 | -0.352 | -0.1259 | No | ||
| 72 | WBSCR16 | 12927 | -0.485 | -0.1461 | No | ||
| 73 | UQCRH | 13079 | -0.527 | -0.1518 | No | ||
| 74 | NFYC | 13127 | -0.539 | -0.1527 | No | ||
| 75 | PRKCSH | 13182 | -0.555 | -0.1539 | No | ||
| 76 | ETV1 | 13308 | -0.590 | -0.1582 | No | ||
| 77 | PSMB5 | 13523 | -0.657 | -0.1665 | No | ||
| 78 | CCND1 | 13583 | -0.671 | -0.1677 | No | ||
| 79 | HOXC4 | 13823 | -0.729 | -0.1769 | No | ||
| 80 | SNRPN | 13906 | -0.754 | -0.1789 | No | ||
| 81 | B3GALT6 | 14020 | -0.785 | -0.1823 | No | ||
| 82 | ATP5A1 | 14097 | -0.802 | -0.1839 | No | ||
| 83 | IER2 | 14151 | -0.817 | -0.1845 | No | ||
| 84 | ZBTB4 | 14269 | -0.843 | -0.1879 | No | ||
| 85 | IRAK1 | 14459 | -0.896 | -0.1945 | No | ||
| 86 | EIF4A1 | 14880 | -1.009 | -0.2114 | No | ||
| 87 | PIGL | 14885 | -1.011 | -0.2092 | No | ||
| 88 | TMEM15 | 15041 | -1.057 | -0.2139 | No | ||
| 89 | TAX1BP3 | 15440 | -1.166 | -0.2295 | No | ||
| 90 | CPEB4 | 15466 | -1.173 | -0.2279 | No | ||
| 91 | RIMS1 | 15889 | -1.281 | -0.2443 | No | ||
| 92 | ARF1 | 15903 | -1.285 | -0.2419 | No | ||
| 93 | BOP1 | 16009 | -1.314 | -0.2436 | No | ||
| 94 | TRIM8 | 16203 | -1.361 | -0.2493 | No | ||
| 95 | NCDN | 16277 | -1.383 | -0.2495 | No | ||
| 96 | MAP3K4 | 16508 | -1.443 | -0.2567 | No | ||
| 97 | CLK2 | 16729 | -1.506 | -0.2633 | No | ||
| 98 | ATP5G2 | 17156 | -1.634 | -0.2790 | No | ||
| 99 | SMYD5 | 17428 | -1.728 | -0.2874 | No | ||
| 100 | PIAS3 | 17585 | -1.780 | -0.2905 | No | ||
| 101 | FZD8 | 17587 | -1.780 | -0.2864 | No | ||
| 102 | RPN1 | 17810 | -1.853 | -0.2923 | Yes | ||
| 103 | PBX3 | 17849 | -1.869 | -0.2897 | Yes | ||
| 104 | HSF1 | 17881 | -1.882 | -0.2867 | Yes | ||
| 105 | TOP3A | 17995 | -1.918 | -0.2874 | Yes | ||
| 106 | PNRC2 | 18026 | -1.929 | -0.2843 | Yes | ||
| 107 | SNX13 | 18051 | -1.942 | -0.2809 | Yes | ||
| 108 | SLC26A6 | 18064 | -1.945 | -0.2769 | Yes | ||
| 109 | RBM3 | 18079 | -1.950 | -0.2730 | Yes | ||
| 110 | TNRC15 | 18179 | -1.987 | -0.2730 | Yes | ||
| 111 | RAB28 | 18228 | -2.005 | -0.2705 | Yes | ||
| 112 | ADK | 18271 | -2.024 | -0.2677 | Yes | ||
| 113 | UBE4B | 18346 | -2.057 | -0.2663 | Yes | ||
| 114 | SF3A1 | 18423 | -2.087 | -0.2650 | Yes | ||
| 115 | EPC1 | 18893 | -2.291 | -0.2812 | Yes | ||
| 116 | RALA | 18965 | -2.321 | -0.2790 | Yes | ||
| 117 | HIC2 | 19020 | -2.347 | -0.2761 | Yes | ||
| 118 | CCDC6 | 19113 | -2.398 | -0.2747 | Yes | ||
| 119 | UBE1 | 19127 | -2.406 | -0.2697 | Yes | ||
| 120 | TBC1D14 | 19140 | -2.412 | -0.2647 | Yes | ||
| 121 | SLC25A19 | 19391 | -2.543 | -0.2702 | Yes | ||
| 122 | PUM1 | 19395 | -2.547 | -0.2644 | Yes | ||
| 123 | BTRC | 19514 | -2.615 | -0.2638 | Yes | ||
| 124 | CHFR | 19588 | -2.652 | -0.2610 | Yes | ||
| 125 | ZFP37 | 19590 | -2.653 | -0.2548 | Yes | ||
| 126 | ALG12 | 19610 | -2.664 | -0.2495 | Yes | ||
| 127 | DDX23 | 19698 | -2.710 | -0.2472 | Yes | ||
| 128 | RPL35A | 19735 | -2.733 | -0.2425 | Yes | ||
| 129 | LPHN1 | 19812 | -2.773 | -0.2395 | Yes | ||
| 130 | GTPBP1 | 19856 | -2.803 | -0.2350 | Yes | ||
| 131 | RNF26 | 19876 | -2.812 | -0.2293 | Yes | ||
| 132 | PTBP1 | 19927 | -2.846 | -0.2250 | Yes | ||
| 133 | RAB22A | 20059 | -2.931 | -0.2242 | Yes | ||
| 134 | STRN4 | 20086 | -2.958 | -0.2185 | Yes | ||
| 135 | ARNT | 20092 | -2.961 | -0.2119 | Yes | ||
| 136 | ASH1L | 20267 | -3.070 | -0.2127 | Yes | ||
| 137 | PPP1R12B | 20296 | -3.097 | -0.2068 | Yes | ||
| 138 | CSNK1A1 | 20494 | -3.270 | -0.2082 | Yes | ||
| 139 | RIF1 | 20509 | -3.284 | -0.2012 | Yes | ||
| 140 | POLR2A | 20527 | -3.291 | -0.1944 | Yes | ||
| 141 | DGCR2 | 20567 | -3.325 | -0.1884 | Yes | ||
| 142 | MLL5 | 20665 | -3.418 | -0.1849 | Yes | ||
| 143 | ENSA | 20788 | -3.553 | -0.1823 | Yes | ||
| 144 | NDRG3 | 20802 | -3.570 | -0.1746 | Yes | ||
| 145 | ATP5H | 20804 | -3.572 | -0.1663 | Yes | ||
| 146 | TAF6 | 20861 | -3.645 | -0.1604 | Yes | ||
| 147 | DZIP1 | 21083 | -3.943 | -0.1614 | Yes | ||
| 148 | DDX6 | 21125 | -4.012 | -0.1539 | Yes | ||
| 149 | ECH1 | 21132 | -4.021 | -0.1449 | Yes | ||
| 150 | ZZEF1 | 21354 | -4.495 | -0.1446 | Yes | ||
| 151 | SMCR8 | 21427 | -4.681 | -0.1370 | Yes | ||
| 152 | UTX | 21431 | -4.694 | -0.1262 | Yes | ||
| 153 | CASC3 | 21458 | -4.756 | -0.1163 | Yes | ||
| 154 | UBE2J2 | 21484 | -4.825 | -0.1063 | Yes | ||
| 155 | AP3D1 | 21496 | -4.863 | -0.0955 | Yes | ||
| 156 | PMF1 | 21528 | -4.995 | -0.0853 | Yes | ||
| 157 | CNOT3 | 21570 | -5.136 | -0.0752 | Yes | ||
| 158 | GIT2 | 21641 | -5.399 | -0.0659 | Yes | ||
| 159 | TSC1 | 21728 | -5.921 | -0.0561 | Yes | ||
| 160 | TIA1 | 21757 | -6.125 | -0.0431 | Yes | ||
| 161 | GBF1 | 21781 | -6.360 | -0.0294 | Yes | ||
| 162 | FOXO3A | 21941 | -15.889 | 0.0003 | Yes |