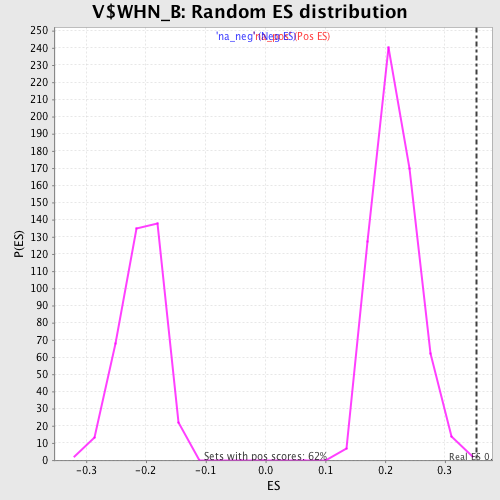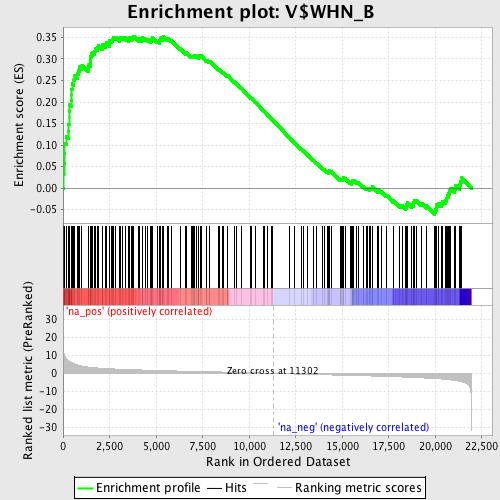
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set01_ATM_minus_versus_SCID |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_pos |
| GeneSet | V$WHN_B |
| Enrichment Score (ES) | 0.35301176 |
| Normalized Enrichment Score (NES) | 1.6278526 |
| Nominal p-value | 0.001607717 |
| FDR q-value | 0.08025459 |
| FWER p-Value | 0.678 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | SLC25A4 | 17 | 14.011 | 0.0329 | Yes | ||
| 2 | YTHDF2 | 48 | 11.012 | 0.0580 | Yes | ||
| 3 | AMD1 | 69 | 10.138 | 0.0815 | Yes | ||
| 4 | CCAR1 | 81 | 9.656 | 0.1042 | Yes | ||
| 5 | GGPS1 | 162 | 8.324 | 0.1205 | Yes | ||
| 6 | SSB | 280 | 6.995 | 0.1320 | Yes | ||
| 7 | ARIH1 | 286 | 6.974 | 0.1485 | Yes | ||
| 8 | UCHL5 | 319 | 6.698 | 0.1631 | Yes | ||
| 9 | DDX5 | 320 | 6.691 | 0.1792 | Yes | ||
| 10 | CXADR | 354 | 6.442 | 0.1932 | Yes | ||
| 11 | MBNL1 | 447 | 5.895 | 0.2032 | Yes | ||
| 12 | HSPD1 | 465 | 5.801 | 0.2163 | Yes | ||
| 13 | SF3B1 | 467 | 5.791 | 0.2302 | Yes | ||
| 14 | ANKRD17 | 486 | 5.653 | 0.2430 | Yes | ||
| 15 | NOLC1 | 581 | 5.207 | 0.2512 | Yes | ||
| 16 | HSPA14 | 632 | 4.998 | 0.2609 | Yes | ||
| 17 | NIN | 776 | 4.612 | 0.2654 | Yes | ||
| 18 | CDC26 | 825 | 4.484 | 0.2740 | Yes | ||
| 19 | JARID2 | 891 | 4.340 | 0.2814 | Yes | ||
| 20 | LEF1 | 1013 | 4.051 | 0.2856 | Yes | ||
| 21 | EXTL2 | 1346 | 3.501 | 0.2788 | Yes | ||
| 22 | MIB1 | 1369 | 3.461 | 0.2861 | Yes | ||
| 23 | STAG2 | 1457 | 3.358 | 0.2902 | Yes | ||
| 24 | PTMA | 1471 | 3.346 | 0.2976 | Yes | ||
| 25 | TOP1 | 1486 | 3.326 | 0.3050 | Yes | ||
| 26 | MYO1E | 1503 | 3.308 | 0.3122 | Yes | ||
| 27 | ADRA2C | 1593 | 3.209 | 0.3158 | Yes | ||
| 28 | VAX1 | 1706 | 3.105 | 0.3182 | Yes | ||
| 29 | NSF | 1740 | 3.077 | 0.3240 | Yes | ||
| 30 | DNAJB4 | 1839 | 2.990 | 0.3267 | Yes | ||
| 31 | EFNB1 | 1900 | 2.939 | 0.3310 | Yes | ||
| 32 | SUPT16H | 2103 | 2.765 | 0.3284 | Yes | ||
| 33 | NPR3 | 2136 | 2.742 | 0.3335 | Yes | ||
| 34 | PFN2 | 2299 | 2.641 | 0.3324 | Yes | ||
| 35 | RNF6 | 2317 | 2.636 | 0.3380 | Yes | ||
| 36 | HOXA3 | 2496 | 2.540 | 0.3359 | Yes | ||
| 37 | SENP2 | 2498 | 2.538 | 0.3420 | Yes | ||
| 38 | ZBTB11 | 2592 | 2.484 | 0.3437 | Yes | ||
| 39 | KLF2 | 2674 | 2.445 | 0.3458 | Yes | ||
| 40 | HOXA7 | 2696 | 2.434 | 0.3507 | Yes | ||
| 41 | SNX9 | 2824 | 2.375 | 0.3506 | Yes | ||
| 42 | NR2E1 | 3053 | 2.277 | 0.3456 | Yes | ||
| 43 | HDAC4 | 3069 | 2.271 | 0.3504 | Yes | ||
| 44 | INSM1 | 3181 | 2.227 | 0.3506 | Yes | ||
| 45 | TAC1 | 3329 | 2.167 | 0.3491 | Yes | ||
| 46 | HOXC6 | 3502 | 2.101 | 0.3462 | Yes | ||
| 47 | ARID4B | 3554 | 2.079 | 0.3489 | Yes | ||
| 48 | SNCB | 3675 | 2.034 | 0.3483 | Yes | ||
| 49 | COL4A5 | 3729 | 2.016 | 0.3507 | Yes | ||
| 50 | GRIN2A | 3784 | 1.997 | 0.3530 | Yes | ||
| 51 | NEUROG3 | 4041 | 1.914 | 0.3459 | No | ||
| 52 | TAF5 | 4094 | 1.897 | 0.3480 | No | ||
| 53 | SOX3 | 4239 | 1.852 | 0.3459 | No | ||
| 54 | PMP22 | 4242 | 1.850 | 0.3502 | No | ||
| 55 | POU4F1 | 4414 | 1.797 | 0.3467 | No | ||
| 56 | ZFHX1B | 4532 | 1.759 | 0.3455 | No | ||
| 57 | NRG1 | 4674 | 1.708 | 0.3432 | No | ||
| 58 | TBX3 | 4755 | 1.681 | 0.3435 | No | ||
| 59 | KLHL13 | 4773 | 1.676 | 0.3468 | No | ||
| 60 | POU3F4 | 4780 | 1.673 | 0.3505 | No | ||
| 61 | LAPTM4A | 5059 | 1.587 | 0.3416 | No | ||
| 62 | BBS5 | 5183 | 1.547 | 0.3396 | No | ||
| 63 | ZCCHC9 | 5187 | 1.545 | 0.3432 | No | ||
| 64 | JUN | 5219 | 1.538 | 0.3455 | No | ||
| 65 | STC2 | 5230 | 1.535 | 0.3487 | No | ||
| 66 | SALL1 | 5345 | 1.505 | 0.3471 | No | ||
| 67 | DLG5 | 5354 | 1.502 | 0.3504 | No | ||
| 68 | H3F3B | 5398 | 1.490 | 0.3520 | No | ||
| 69 | PHF5A | 5598 | 1.435 | 0.3463 | No | ||
| 70 | TRIB1 | 5655 | 1.422 | 0.3471 | No | ||
| 71 | PHF15 | 5799 | 1.382 | 0.3439 | No | ||
| 72 | GRIN2B | 6287 | 1.264 | 0.3245 | No | ||
| 73 | SPRY2 | 6601 | 1.196 | 0.3130 | No | ||
| 74 | JUP | 6615 | 1.192 | 0.3153 | No | ||
| 75 | FOXA1 | 6873 | 1.128 | 0.3062 | No | ||
| 76 | HOXB9 | 6942 | 1.112 | 0.3058 | No | ||
| 77 | SDCBP2 | 7012 | 1.096 | 0.3052 | No | ||
| 78 | DNAH9 | 7035 | 1.090 | 0.3068 | No | ||
| 79 | NMNAT2 | 7057 | 1.085 | 0.3085 | No | ||
| 80 | KCNS3 | 7178 | 1.056 | 0.3055 | No | ||
| 81 | BDNF | 7264 | 1.037 | 0.3041 | No | ||
| 82 | HYDIN | 7283 | 1.033 | 0.3058 | No | ||
| 83 | KCTD4 | 7296 | 1.030 | 0.3077 | No | ||
| 84 | JPH2 | 7399 | 1.005 | 0.3054 | No | ||
| 85 | EAF1 | 7416 | 1.002 | 0.3071 | No | ||
| 86 | ALK | 7696 | 0.940 | 0.2965 | No | ||
| 87 | TOB2 | 7728 | 0.934 | 0.2974 | No | ||
| 88 | MRPL12 | 7870 | 0.902 | 0.2930 | No | ||
| 89 | PRPF4 | 7888 | 0.897 | 0.2944 | No | ||
| 90 | MYH2 | 8370 | 0.789 | 0.2742 | No | ||
| 91 | NUDT11 | 8397 | 0.784 | 0.2749 | No | ||
| 92 | POU3F3 | 8563 | 0.744 | 0.2691 | No | ||
| 93 | SLC30A7 | 8605 | 0.733 | 0.2690 | No | ||
| 94 | ZBTB12 | 8824 | 0.681 | 0.2606 | No | ||
| 95 | SFN | 8858 | 0.672 | 0.2607 | No | ||
| 96 | ZNF385 | 9184 | 0.597 | 0.2472 | No | ||
| 97 | HOXA9 | 9294 | 0.572 | 0.2436 | No | ||
| 98 | FSTL5 | 9582 | 0.499 | 0.2316 | No | ||
| 99 | CAPN12 | 10093 | 0.357 | 0.2090 | No | ||
| 100 | WNT1 | 10135 | 0.345 | 0.2080 | No | ||
| 101 | SIX1 | 10334 | 0.291 | 0.1996 | No | ||
| 102 | DUSP14 | 10754 | 0.169 | 0.1808 | No | ||
| 103 | NKX2-2 | 10827 | 0.145 | 0.1778 | No | ||
| 104 | TBX6 | 11008 | 0.086 | 0.1697 | No | ||
| 105 | IRX5 | 11189 | 0.033 | 0.1615 | No | ||
| 106 | RAB11A | 11252 | 0.012 | 0.1587 | No | ||
| 107 | CALM2 | 12170 | -0.266 | 0.1172 | No | ||
| 108 | WNT3 | 12450 | -0.349 | 0.1053 | No | ||
| 109 | SYT4 | 12793 | -0.449 | 0.0906 | No | ||
| 110 | FAP | 12903 | -0.478 | 0.0868 | No | ||
| 111 | GABRG2 | 13116 | -0.536 | 0.0783 | No | ||
| 112 | BANP | 13442 | -0.632 | 0.0649 | No | ||
| 113 | BHLHB2 | 13620 | -0.677 | 0.0584 | No | ||
| 114 | ICA1 | 13931 | -0.760 | 0.0460 | No | ||
| 115 | ELAVL3 | 14026 | -0.786 | 0.0436 | No | ||
| 116 | RPL41 | 14180 | -0.824 | 0.0385 | No | ||
| 117 | ONECUT1 | 14261 | -0.841 | 0.0369 | No | ||
| 118 | ZBTB4 | 14269 | -0.843 | 0.0386 | No | ||
| 119 | EGR3 | 14289 | -0.850 | 0.0397 | No | ||
| 120 | MLLT7 | 14309 | -0.856 | 0.0409 | No | ||
| 121 | CTCF | 14413 | -0.882 | 0.0383 | No | ||
| 122 | EIF4A1 | 14880 | -1.009 | 0.0193 | No | ||
| 123 | LRP1B | 14891 | -1.013 | 0.0213 | No | ||
| 124 | JAG1 | 14969 | -1.039 | 0.0203 | No | ||
| 125 | COL4A6 | 15024 | -1.053 | 0.0203 | No | ||
| 126 | TMEM15 | 15041 | -1.057 | 0.0221 | No | ||
| 127 | GRIA3 | 15046 | -1.059 | 0.0245 | No | ||
| 128 | CDC14B | 15155 | -1.088 | 0.0221 | No | ||
| 129 | RFX4 | 15426 | -1.162 | 0.0125 | No | ||
| 130 | PRSS16 | 15494 | -1.181 | 0.0123 | No | ||
| 131 | NF1 | 15523 | -1.189 | 0.0139 | No | ||
| 132 | LMO3 | 15539 | -1.191 | 0.0160 | No | ||
| 133 | ESRRG | 15568 | -1.198 | 0.0176 | No | ||
| 134 | TOB1 | 15629 | -1.215 | 0.0178 | No | ||
| 135 | HOXB8 | 15788 | -1.257 | 0.0136 | No | ||
| 136 | RGL1 | 15856 | -1.275 | 0.0136 | No | ||
| 137 | GALK2 | 16136 | -1.344 | 0.0040 | No | ||
| 138 | CHST11 | 16296 | -1.389 | 0.0000 | No | ||
| 139 | PCDH7 | 16364 | -1.406 | 0.0003 | No | ||
| 140 | SNF1LK | 16473 | -1.434 | -0.0012 | No | ||
| 141 | ARPC5 | 16517 | -1.445 | 0.0003 | No | ||
| 142 | HAPLN1 | 16601 | -1.469 | 0.0000 | No | ||
| 143 | WNT9B | 16608 | -1.471 | 0.0033 | No | ||
| 144 | EML2 | 16903 | -1.553 | -0.0065 | No | ||
| 145 | SLITRK5 | 16929 | -1.563 | -0.0039 | No | ||
| 146 | PAPPA | 17083 | -1.611 | -0.0070 | No | ||
| 147 | DLL4 | 17365 | -1.707 | -0.0158 | No | ||
| 148 | FOXP2 | 17726 | -1.828 | -0.0280 | No | ||
| 149 | PPARGC1A | 18084 | -1.952 | -0.0397 | No | ||
| 150 | OPHN1 | 18218 | -2.002 | -0.0410 | No | ||
| 151 | ID2 | 18403 | -2.081 | -0.0444 | No | ||
| 152 | HERC1 | 18446 | -2.096 | -0.0413 | No | ||
| 153 | JUNB | 18468 | -2.107 | -0.0372 | No | ||
| 154 | PRDM10 | 18478 | -2.111 | -0.0326 | No | ||
| 155 | ELOVL6 | 18737 | -2.219 | -0.0391 | No | ||
| 156 | DDX17 | 18811 | -2.253 | -0.0370 | No | ||
| 157 | NUDT10 | 18825 | -2.259 | -0.0322 | No | ||
| 158 | SLC20A1 | 18858 | -2.271 | -0.0282 | No | ||
| 159 | PPP3CA | 18969 | -2.322 | -0.0277 | No | ||
| 160 | NFKB2 | 19250 | -2.474 | -0.0346 | No | ||
| 161 | BTRC | 19514 | -2.615 | -0.0404 | No | ||
| 162 | NDOR1 | 19967 | -2.870 | -0.0542 | No | ||
| 163 | NRK | 20000 | -2.898 | -0.0487 | No | ||
| 164 | ACO2 | 20054 | -2.927 | -0.0441 | No | ||
| 165 | HPCAL1 | 20078 | -2.948 | -0.0381 | No | ||
| 166 | SFRS12 | 20190 | -3.022 | -0.0359 | No | ||
| 167 | SUMO1 | 20323 | -3.118 | -0.0345 | No | ||
| 168 | SYNJ1 | 20392 | -3.165 | -0.0300 | No | ||
| 169 | POLR2A | 20527 | -3.291 | -0.0282 | No | ||
| 170 | CRYL1 | 20606 | -3.363 | -0.0237 | No | ||
| 171 | THRA | 20628 | -3.380 | -0.0166 | No | ||
| 172 | CAMK4 | 20719 | -3.468 | -0.0124 | No | ||
| 173 | NR4A3 | 20751 | -3.509 | -0.0054 | No | ||
| 174 | TOMM40 | 20829 | -3.600 | -0.0002 | No | ||
| 175 | PAX3 | 21049 | -3.891 | -0.0009 | No | ||
| 176 | H2AFZ | 21106 | -3.980 | 0.0061 | No | ||
| 177 | HSPE1 | 21303 | -4.365 | 0.0075 | No | ||
| 178 | MTMR4 | 21350 | -4.481 | 0.0162 | No | ||
| 179 | DHX30 | 21418 | -4.645 | 0.0243 | No |