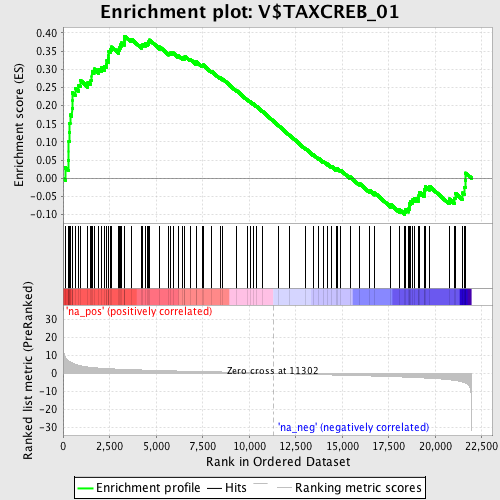
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set01_ATM_minus_versus_SCID |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_pos |
| GeneSet | V$TAXCREB_01 |
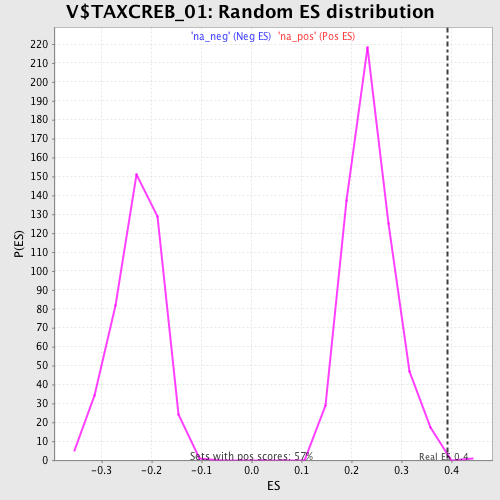
| Enrichment Score (ES) | 0.39137614 |
| Normalized Enrichment Score (NES) | 1.6486317 |
| Nominal p-value | 0.0017421603 |
| FDR q-value | 0.07573061 |
| FWER p-Value | 0.596 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | GSPT1 | 122 | 8.818 | 0.0291 | Yes | ||
| 2 | ARIH1 | 286 | 6.974 | 0.0491 | Yes | ||
| 3 | MBNL2 | 310 | 6.749 | 0.0746 | Yes | ||
| 4 | SLC25A5 | 311 | 6.743 | 0.1011 | Yes | ||
| 5 | NINJ1 | 333 | 6.588 | 0.1261 | Yes | ||
| 6 | EIF4A2 | 356 | 6.429 | 0.1504 | Yes | ||
| 7 | USP14 | 373 | 6.316 | 0.1745 | Yes | ||
| 8 | WHSC1L1 | 477 | 5.742 | 0.1923 | Yes | ||
| 9 | DLAT | 494 | 5.611 | 0.2137 | Yes | ||
| 10 | LUC7L2 | 500 | 5.580 | 0.2354 | Yes | ||
| 11 | APPBP2 | 669 | 4.907 | 0.2470 | Yes | ||
| 12 | SFRS1 | 843 | 4.448 | 0.2566 | Yes | ||
| 13 | MLF2 | 923 | 4.240 | 0.2697 | Yes | ||
| 14 | KDELR3 | 1333 | 3.514 | 0.2648 | Yes | ||
| 15 | POLDIP3 | 1494 | 3.319 | 0.2705 | Yes | ||
| 16 | NLK | 1550 | 3.262 | 0.2808 | Yes | ||
| 17 | CYLD | 1552 | 3.260 | 0.2936 | Yes | ||
| 18 | PLK4 | 1661 | 3.143 | 0.3010 | Yes | ||
| 19 | PPP2CB | 1920 | 2.923 | 0.3007 | Yes | ||
| 20 | OSR1 | 2071 | 2.789 | 0.3048 | Yes | ||
| 21 | ALS2CR13 | 2234 | 2.681 | 0.3079 | Yes | ||
| 22 | ATF1 | 2314 | 2.637 | 0.3147 | Yes | ||
| 23 | SSTR3 | 2353 | 2.613 | 0.3232 | Yes | ||
| 24 | XPR1 | 2434 | 2.569 | 0.3297 | Yes | ||
| 25 | DLST | 2453 | 2.561 | 0.3389 | Yes | ||
| 26 | FBXL11 | 2459 | 2.558 | 0.3488 | Yes | ||
| 27 | ARRB2 | 2541 | 2.515 | 0.3550 | Yes | ||
| 28 | NTRK2 | 2611 | 2.474 | 0.3615 | Yes | ||
| 29 | PAK1 | 2989 | 2.306 | 0.3533 | Yes | ||
| 30 | GTF2A1 | 3046 | 2.279 | 0.3597 | Yes | ||
| 31 | CLSTN3 | 3079 | 2.267 | 0.3672 | Yes | ||
| 32 | RING1 | 3154 | 2.238 | 0.3726 | Yes | ||
| 33 | CDC42 | 3285 | 2.187 | 0.3753 | Yes | ||
| 34 | PCF11 | 3289 | 2.185 | 0.3837 | Yes | ||
| 35 | BNIP2 | 3310 | 2.176 | 0.3914 | Yes | ||
| 36 | GATA1 | 3662 | 2.039 | 0.3833 | No | ||
| 37 | TAF11 | 4227 | 1.855 | 0.3648 | No | ||
| 38 | MARCKS | 4285 | 1.832 | 0.3694 | No | ||
| 39 | ARMCX3 | 4407 | 1.798 | 0.3709 | No | ||
| 40 | GPM6B | 4530 | 1.761 | 0.3723 | No | ||
| 41 | FOSB | 4608 | 1.729 | 0.3756 | No | ||
| 42 | HS6ST2 | 4643 | 1.715 | 0.3808 | No | ||
| 43 | NR2F1 | 5195 | 1.544 | 0.3616 | No | ||
| 44 | PEG3 | 5684 | 1.413 | 0.3448 | No | ||
| 45 | SIX3 | 5791 | 1.384 | 0.3454 | No | ||
| 46 | CD2AP | 5915 | 1.352 | 0.3451 | No | ||
| 47 | CCND2 | 6194 | 1.286 | 0.3374 | No | ||
| 48 | GEM | 6426 | 1.234 | 0.3317 | No | ||
| 49 | GPC6 | 6507 | 1.216 | 0.3328 | No | ||
| 50 | PNRC1 | 6547 | 1.207 | 0.3358 | No | ||
| 51 | CDK5R2 | 6824 | 1.140 | 0.3277 | No | ||
| 52 | HOXB1 | 7146 | 1.063 | 0.3171 | No | ||
| 53 | IRX4 | 7159 | 1.060 | 0.3208 | No | ||
| 54 | KLF13 | 7470 | 0.991 | 0.3105 | No | ||
| 55 | RAB37 | 7517 | 0.983 | 0.3122 | No | ||
| 56 | SCAMP5 | 7996 | 0.872 | 0.2938 | No | ||
| 57 | SMARCD1 | 8437 | 0.772 | 0.2767 | No | ||
| 58 | FOS | 8574 | 0.740 | 0.2734 | No | ||
| 59 | TAGLN2 | 9342 | 0.561 | 0.2405 | No | ||
| 60 | MAP1LC3A | 9925 | 0.403 | 0.2154 | No | ||
| 61 | TUBA2 | 10063 | 0.366 | 0.2106 | No | ||
| 62 | GPR3 | 10203 | 0.326 | 0.2055 | No | ||
| 63 | PTPRU | 10385 | 0.275 | 0.1983 | No | ||
| 64 | SLC30A5 | 10688 | 0.189 | 0.1852 | No | ||
| 65 | ZIM2 | 11553 | -0.085 | 0.1460 | No | ||
| 66 | CALM2 | 12170 | -0.266 | 0.1188 | No | ||
| 67 | DUSP1 | 13018 | -0.508 | 0.0821 | No | ||
| 68 | RFX5 | 13427 | -0.629 | 0.0659 | No | ||
| 69 | DLX5 | 13742 | -0.710 | 0.0543 | No | ||
| 70 | HOXA10 | 13989 | -0.775 | 0.0461 | No | ||
| 71 | MAFF | 14202 | -0.827 | 0.0396 | No | ||
| 72 | CTCF | 14413 | -0.882 | 0.0335 | No | ||
| 73 | HAS1 | 14705 | -0.959 | 0.0239 | No | ||
| 74 | VGF | 14726 | -0.965 | 0.0268 | No | ||
| 75 | TRIM39 | 14918 | -1.026 | 0.0221 | No | ||
| 76 | HHIP | 15416 | -1.157 | 0.0039 | No | ||
| 77 | CEBPB | 15911 | -1.287 | -0.0136 | No | ||
| 78 | SNF1LK | 16473 | -1.434 | -0.0337 | No | ||
| 79 | NOL4 | 16745 | -1.511 | -0.0402 | No | ||
| 80 | SDHB | 17604 | -1.785 | -0.0724 | No | ||
| 81 | SOX10 | 18083 | -1.952 | -0.0866 | No | ||
| 82 | OTUB2 | 18368 | -2.065 | -0.0915 | No | ||
| 83 | VEGF | 18410 | -2.082 | -0.0852 | No | ||
| 84 | GPR158 | 18578 | -2.148 | -0.0844 | No | ||
| 85 | LMO4 | 18605 | -2.158 | -0.0771 | No | ||
| 86 | VAMP2 | 18615 | -2.163 | -0.0690 | No | ||
| 87 | OSBP | 18675 | -2.191 | -0.0630 | No | ||
| 88 | FGF14 | 18792 | -2.246 | -0.0595 | No | ||
| 89 | MESDC1 | 18881 | -2.284 | -0.0546 | No | ||
| 90 | BRAF | 19077 | -2.378 | -0.0541 | No | ||
| 91 | BAHD1 | 19118 | -2.400 | -0.0465 | No | ||
| 92 | RPS27 | 19139 | -2.412 | -0.0379 | No | ||
| 93 | CDKN1B | 19399 | -2.549 | -0.0398 | No | ||
| 94 | G6PC | 19423 | -2.560 | -0.0308 | No | ||
| 95 | SMARCA5 | 19457 | -2.580 | -0.0221 | No | ||
| 96 | PLCB3 | 19683 | -2.699 | -0.0218 | No | ||
| 97 | NR4A3 | 20751 | -3.509 | -0.0568 | No | ||
| 98 | ASXL1 | 21029 | -3.860 | -0.0543 | No | ||
| 99 | JMJD1B | 21076 | -3.929 | -0.0410 | No | ||
| 100 | TMEM1 | 21475 | -4.802 | -0.0403 | No | ||
| 101 | CAMK2D | 21559 | -5.093 | -0.0241 | No | ||
| 102 | PHF12 | 21607 | -5.257 | -0.0055 | No | ||
| 103 | PAFAH1B1 | 21628 | -5.352 | 0.0146 | No |