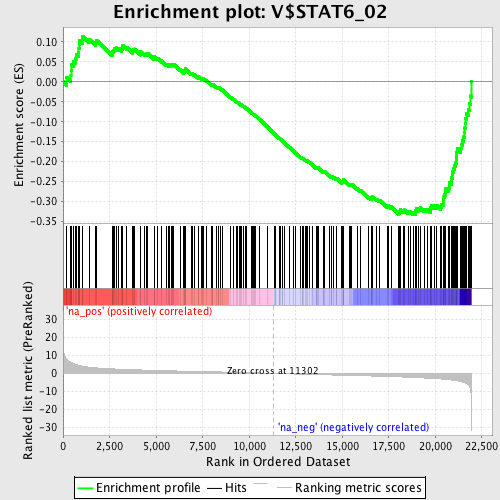
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set01_ATM_minus_versus_SCID |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_neg |
| GeneSet | V$STAT6_02 |
| Enrichment Score (ES) | -0.33221385 |
| Normalized Enrichment Score (NES) | -1.6340677 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.55638826 |
| FWER p-Value | 0.465 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | POLR1D | 185 | 7.972 | 0.0108 | No | ||
| 2 | RNF138 | 421 | 6.052 | 0.0147 | No | ||
| 3 | FEM1C | 425 | 6.037 | 0.0293 | No | ||
| 4 | MBNL1 | 447 | 5.895 | 0.0426 | No | ||
| 5 | IL7R | 551 | 5.343 | 0.0508 | No | ||
| 6 | MCTS1 | 643 | 4.963 | 0.0587 | No | ||
| 7 | SLC31A1 | 695 | 4.828 | 0.0681 | No | ||
| 8 | ZNRF2 | 808 | 4.536 | 0.0739 | No | ||
| 9 | TANK | 849 | 4.434 | 0.0829 | No | ||
| 10 | SRRM1 | 864 | 4.408 | 0.0929 | No | ||
| 11 | DNAJA2 | 878 | 4.367 | 0.1029 | No | ||
| 12 | CENTB2 | 1018 | 4.047 | 0.1064 | No | ||
| 13 | SPI1 | 1054 | 3.971 | 0.1144 | No | ||
| 14 | CRK | 1421 | 3.408 | 0.1058 | No | ||
| 15 | HMGA1 | 1732 | 3.086 | 0.0991 | No | ||
| 16 | EGFL7 | 1783 | 3.044 | 0.1042 | No | ||
| 17 | TRIM25 | 2632 | 2.463 | 0.0712 | No | ||
| 18 | PGM2L1 | 2664 | 2.450 | 0.0757 | No | ||
| 19 | JARID1C | 2720 | 2.424 | 0.0790 | No | ||
| 20 | VAV3 | 2767 | 2.404 | 0.0828 | No | ||
| 21 | ANKRD28 | 2847 | 2.368 | 0.0849 | No | ||
| 22 | CITED4 | 2978 | 2.311 | 0.0845 | No | ||
| 23 | ITGA8 | 3155 | 2.238 | 0.0819 | No | ||
| 24 | ZF | 3177 | 2.229 | 0.0863 | No | ||
| 25 | DLC1 | 3209 | 2.217 | 0.0903 | No | ||
| 26 | HIF3A | 3398 | 2.141 | 0.0868 | No | ||
| 27 | DHH | 3724 | 2.017 | 0.0768 | No | ||
| 28 | EPHA7 | 3763 | 2.004 | 0.0799 | No | ||
| 29 | LY75 | 3820 | 1.985 | 0.0821 | No | ||
| 30 | A2BP1 | 4149 | 1.881 | 0.0716 | No | ||
| 31 | LUC7L | 4153 | 1.879 | 0.0760 | No | ||
| 32 | HSD11B1 | 4398 | 1.800 | 0.0692 | No | ||
| 33 | ZRANB1 | 4480 | 1.775 | 0.0698 | No | ||
| 34 | ZFHX1B | 4532 | 1.759 | 0.0717 | No | ||
| 35 | MPO | 4889 | 1.640 | 0.0593 | No | ||
| 36 | LALBA | 4894 | 1.638 | 0.0631 | No | ||
| 37 | PDGFRB | 5066 | 1.584 | 0.0591 | No | ||
| 38 | CHD6 | 5283 | 1.522 | 0.0529 | No | ||
| 39 | MTUS1 | 5546 | 1.447 | 0.0443 | No | ||
| 40 | LAG3 | 5675 | 1.414 | 0.0419 | No | ||
| 41 | ACBD6 | 5734 | 1.399 | 0.0426 | No | ||
| 42 | PHF15 | 5799 | 1.382 | 0.0430 | No | ||
| 43 | CENTA1 | 5877 | 1.362 | 0.0428 | No | ||
| 44 | CD2AP | 5915 | 1.352 | 0.0444 | No | ||
| 45 | GRIN2B | 6287 | 1.264 | 0.0304 | No | ||
| 46 | FLNC | 6494 | 1.218 | 0.0239 | No | ||
| 47 | ERG | 6504 | 1.216 | 0.0264 | No | ||
| 48 | PDZRN4 | 6539 | 1.210 | 0.0278 | No | ||
| 49 | ADCYAP1 | 6563 | 1.204 | 0.0297 | No | ||
| 50 | PLCB2 | 6566 | 1.204 | 0.0325 | No | ||
| 51 | SESTD1 | 6880 | 1.127 | 0.0208 | No | ||
| 52 | HOXB9 | 6942 | 1.112 | 0.0207 | No | ||
| 53 | NMNAT2 | 7057 | 1.085 | 0.0181 | No | ||
| 54 | BDNF | 7264 | 1.037 | 0.0112 | No | ||
| 55 | BMP2 | 7273 | 1.035 | 0.0133 | No | ||
| 56 | FLT1 | 7430 | 0.998 | 0.0086 | No | ||
| 57 | CSF3R | 7514 | 0.983 | 0.0071 | No | ||
| 58 | LAT | 7545 | 0.978 | 0.0081 | No | ||
| 59 | NOSTRIN | 7697 | 0.940 | 0.0035 | No | ||
| 60 | USP2 | 7960 | 0.881 | -0.0064 | No | ||
| 61 | LTC4S | 8034 | 0.862 | -0.0077 | No | ||
| 62 | RHOJ | 8045 | 0.859 | -0.0061 | No | ||
| 63 | EPHA2 | 8247 | 0.813 | -0.0133 | No | ||
| 64 | WNT5A | 8347 | 0.794 | -0.0159 | No | ||
| 65 | PSTPIP2 | 8361 | 0.792 | -0.0146 | No | ||
| 66 | ARV1 | 8452 | 0.770 | -0.0169 | No | ||
| 67 | POU3F3 | 8563 | 0.744 | -0.0201 | No | ||
| 68 | GREM1 | 8995 | 0.640 | -0.0384 | No | ||
| 69 | NOTCH4 | 9146 | 0.605 | -0.0438 | No | ||
| 70 | AKT1S1 | 9328 | 0.564 | -0.0508 | No | ||
| 71 | PKN3 | 9359 | 0.557 | -0.0508 | No | ||
| 72 | EDN1 | 9472 | 0.531 | -0.0547 | No | ||
| 73 | SCGB3A1 | 9557 | 0.504 | -0.0573 | No | ||
| 74 | SDCBP | 9591 | 0.495 | -0.0576 | No | ||
| 75 | RNF128 | 9692 | 0.466 | -0.0611 | No | ||
| 76 | HHEX | 9811 | 0.435 | -0.0654 | No | ||
| 77 | MIST | 9874 | 0.416 | -0.0673 | No | ||
| 78 | ZNF289 | 10101 | 0.354 | -0.0768 | No | ||
| 79 | SPDEF | 10156 | 0.337 | -0.0785 | No | ||
| 80 | RUNX3 | 10254 | 0.313 | -0.0822 | No | ||
| 81 | RAB39 | 10259 | 0.312 | -0.0816 | No | ||
| 82 | PAX9 | 10350 | 0.287 | -0.0850 | No | ||
| 83 | DCTN2 | 10568 | 0.223 | -0.0945 | No | ||
| 84 | CTNNB1 | 11002 | 0.088 | -0.1142 | No | ||
| 85 | CYB561D2 | 11345 | -0.014 | -0.1299 | No | ||
| 86 | TLL1 | 11397 | -0.031 | -0.1321 | No | ||
| 87 | GNGT2 | 11430 | -0.045 | -0.1335 | No | ||
| 88 | HGFAC | 11636 | -0.106 | -0.1427 | No | ||
| 89 | TWIST1 | 11637 | -0.106 | -0.1424 | No | ||
| 90 | STAB1 | 11690 | -0.124 | -0.1445 | No | ||
| 91 | SNX8 | 11772 | -0.147 | -0.1478 | No | ||
| 92 | GSC | 11814 | -0.162 | -0.1493 | No | ||
| 93 | ADSL | 11915 | -0.188 | -0.1535 | No | ||
| 94 | TFEC | 12154 | -0.261 | -0.1638 | No | ||
| 95 | LRRC19 | 12391 | -0.327 | -0.1738 | No | ||
| 96 | CPA3 | 12489 | -0.362 | -0.1774 | No | ||
| 97 | HNRPUL1 | 12768 | -0.442 | -0.1891 | No | ||
| 98 | HOXA2 | 12841 | -0.462 | -0.1913 | No | ||
| 99 | TCF15 | 12861 | -0.465 | -0.1911 | No | ||
| 100 | TOM1L1 | 12932 | -0.486 | -0.1931 | No | ||
| 101 | GPS2 | 13035 | -0.514 | -0.1965 | No | ||
| 102 | SGK | 13093 | -0.530 | -0.1979 | No | ||
| 103 | SLIT3 | 13105 | -0.533 | -0.1971 | No | ||
| 104 | RAB1B | 13233 | -0.569 | -0.2015 | No | ||
| 105 | NKIRAS2 | 13419 | -0.627 | -0.2085 | No | ||
| 106 | CASKIN2 | 13589 | -0.672 | -0.2147 | No | ||
| 107 | ZDHHC8 | 13634 | -0.680 | -0.2150 | No | ||
| 108 | PIM2 | 13650 | -0.683 | -0.2141 | No | ||
| 109 | SLC35C1 | 13712 | -0.703 | -0.2152 | No | ||
| 110 | HOXA10 | 13989 | -0.775 | -0.2260 | No | ||
| 111 | COX17 | 13997 | -0.777 | -0.2244 | No | ||
| 112 | ARID1A | 14066 | -0.796 | -0.2256 | No | ||
| 113 | TNFSF13 | 14306 | -0.855 | -0.2345 | No | ||
| 114 | CTCF | 14413 | -0.882 | -0.2372 | No | ||
| 115 | CNN2 | 14440 | -0.889 | -0.2363 | No | ||
| 116 | VASP | 14554 | -0.919 | -0.2392 | No | ||
| 117 | FEV | 14678 | -0.953 | -0.2426 | No | ||
| 118 | MRPL24 | 14711 | -0.961 | -0.2417 | No | ||
| 119 | CRYGB | 14970 | -1.039 | -0.2510 | No | ||
| 120 | LRP2 | 15013 | -1.049 | -0.2504 | No | ||
| 121 | STK4 | 15042 | -1.057 | -0.2491 | No | ||
| 122 | HIVEP1 | 15050 | -1.060 | -0.2469 | No | ||
| 123 | ERF | 15068 | -1.064 | -0.2451 | No | ||
| 124 | HTR3B | 15389 | -1.151 | -0.2570 | No | ||
| 125 | CPEB4 | 15466 | -1.173 | -0.2577 | No | ||
| 126 | ASPN | 15501 | -1.182 | -0.2564 | No | ||
| 127 | RANBP9 | 15801 | -1.260 | -0.2670 | No | ||
| 128 | GPX1 | 15966 | -1.302 | -0.2714 | No | ||
| 129 | FGF13 | 16416 | -1.420 | -0.2886 | No | ||
| 130 | PDE4D | 16569 | -1.458 | -0.2921 | No | ||
| 131 | GZMK | 16595 | -1.468 | -0.2896 | No | ||
| 132 | CA3 | 16639 | -1.479 | -0.2880 | No | ||
| 133 | RGS3 | 16853 | -1.540 | -0.2941 | No | ||
| 134 | EGF | 16992 | -1.583 | -0.2966 | No | ||
| 135 | CACNG3 | 17412 | -1.724 | -0.3117 | No | ||
| 136 | CITED2 | 17464 | -1.741 | -0.3098 | No | ||
| 137 | CLOCK | 17634 | -1.796 | -0.3132 | No | ||
| 138 | SLC43A1 | 18028 | -1.929 | -0.3266 | No | ||
| 139 | ANGPTL1 | 18093 | -1.956 | -0.3248 | No | ||
| 140 | PAX8 | 18103 | -1.958 | -0.3204 | No | ||
| 141 | DOCK4 | 18272 | -2.024 | -0.3232 | No | ||
| 142 | CSF2 | 18330 | -2.052 | -0.3209 | No | ||
| 143 | RIN2 | 18547 | -2.138 | -0.3256 | No | ||
| 144 | HOXB4 | 18643 | -2.180 | -0.3247 | No | ||
| 145 | LNX2 | 18808 | -2.253 | -0.3267 | Yes | ||
| 146 | RBBP5 | 18919 | -2.302 | -0.3262 | Yes | ||
| 147 | SLC38A2 | 18943 | -2.312 | -0.3217 | Yes | ||
| 148 | PDLIM2 | 18973 | -2.324 | -0.3173 | Yes | ||
| 149 | TRERF1 | 19119 | -2.401 | -0.3182 | Yes | ||
| 150 | TBC1D17 | 19195 | -2.443 | -0.3157 | Yes | ||
| 151 | PUM1 | 19395 | -2.547 | -0.3187 | Yes | ||
| 152 | MEF2C | 19552 | -2.634 | -0.3194 | Yes | ||
| 153 | VAMP3 | 19721 | -2.724 | -0.3206 | Yes | ||
| 154 | MAP4K2 | 19737 | -2.734 | -0.3146 | Yes | ||
| 155 | BZRAP1 | 19789 | -2.758 | -0.3103 | Yes | ||
| 156 | ABI3 | 19951 | -2.857 | -0.3107 | Yes | ||
| 157 | VPS39 | 20077 | -2.946 | -0.3093 | Yes | ||
| 158 | PRKCB1 | 20298 | -3.099 | -0.3119 | Yes | ||
| 159 | SERPINI1 | 20341 | -3.130 | -0.3062 | Yes | ||
| 160 | TTC13 | 20415 | -3.182 | -0.3019 | Yes | ||
| 161 | PDCD10 | 20445 | -3.216 | -0.2954 | Yes | ||
| 162 | ATP5E | 20463 | -3.238 | -0.2883 | Yes | ||
| 163 | CMYA5 | 20480 | -3.253 | -0.2812 | Yes | ||
| 164 | TRAF3 | 20536 | -3.298 | -0.2757 | Yes | ||
| 165 | STRC | 20547 | -3.305 | -0.2681 | Yes | ||
| 166 | DNAJC7 | 20690 | -3.440 | -0.2663 | Yes | ||
| 167 | CBLB | 20738 | -3.491 | -0.2600 | Yes | ||
| 168 | TRPS1 | 20749 | -3.507 | -0.2519 | Yes | ||
| 169 | SATB1 | 20856 | -3.642 | -0.2480 | Yes | ||
| 170 | SEMA4B | 20883 | -3.663 | -0.2403 | Yes | ||
| 171 | DOCK11 | 20905 | -3.691 | -0.2323 | Yes | ||
| 172 | TUSC4 | 20917 | -3.706 | -0.2238 | Yes | ||
| 173 | CAMK1D | 20984 | -3.794 | -0.2176 | Yes | ||
| 174 | SIPA1 | 21017 | -3.843 | -0.2097 | Yes | ||
| 175 | ITPR3 | 21081 | -3.940 | -0.2031 | Yes | ||
| 176 | SYTL1 | 21120 | -4.007 | -0.1951 | Yes | ||
| 177 | FBXW11 | 21133 | -4.021 | -0.1859 | Yes | ||
| 178 | STAT5A | 21151 | -4.058 | -0.1768 | Yes | ||
| 179 | GNB2 | 21165 | -4.080 | -0.1675 | Yes | ||
| 180 | GPHN | 21369 | -4.532 | -0.1658 | Yes | ||
| 181 | SAP130 | 21410 | -4.616 | -0.1565 | Yes | ||
| 182 | MAP3K3 | 21465 | -4.765 | -0.1474 | Yes | ||
| 183 | KCNQ1 | 21527 | -4.987 | -0.1381 | Yes | ||
| 184 | NFAT5 | 21578 | -5.169 | -0.1278 | Yes | ||
| 185 | PITPNC1 | 21586 | -5.198 | -0.1155 | Yes | ||
| 186 | PHF12 | 21607 | -5.257 | -0.1037 | Yes | ||
| 187 | FLI1 | 21627 | -5.352 | -0.0916 | Yes | ||
| 188 | PSCD4 | 21666 | -5.511 | -0.0800 | Yes | ||
| 189 | LCK | 21782 | -6.376 | -0.0698 | Yes | ||
| 190 | PPP2R5C | 21843 | -7.340 | -0.0547 | Yes | ||
| 191 | SLC1A5 | 21879 | -8.578 | -0.0355 | Yes | ||
| 192 | FOXO3A | 21941 | -15.889 | 0.0003 | Yes |