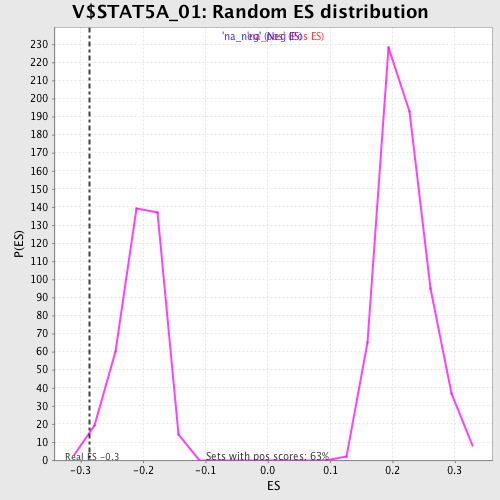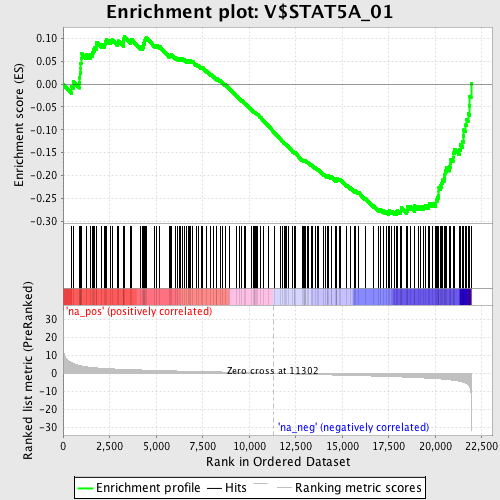
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set01_ATM_minus_versus_SCID |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_neg |
| GeneSet | V$STAT5A_01 |
| Enrichment Score (ES) | -0.28548 |
| Normalized Enrichment Score (NES) | -1.3957227 |
| Nominal p-value | 0.018817205 |
| FDR q-value | 0.42696077 |
| FWER p-Value | 0.997 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | ETV5 | 461 | 5.827 | -0.0052 | No | ||
| 2 | BCLAF1 | 552 | 5.339 | 0.0053 | No | ||
| 3 | HNRPR | 858 | 4.419 | 0.0035 | No | ||
| 4 | CCL2 | 900 | 4.322 | 0.0134 | No | ||
| 5 | ATP8A2 | 907 | 4.285 | 0.0249 | No | ||
| 6 | ATP2A2 | 947 | 4.197 | 0.0346 | No | ||
| 7 | NDST1 | 951 | 4.190 | 0.0460 | No | ||
| 8 | YWHAQ | 965 | 4.155 | 0.0568 | No | ||
| 9 | COL4A3BP | 981 | 4.127 | 0.0675 | No | ||
| 10 | ATP11C | 1237 | 3.662 | 0.0658 | No | ||
| 11 | NRXN3 | 1468 | 3.349 | 0.0644 | No | ||
| 12 | UBR1 | 1569 | 3.234 | 0.0687 | No | ||
| 13 | SOD1 | 1609 | 3.197 | 0.0757 | No | ||
| 14 | VAX1 | 1706 | 3.105 | 0.0798 | No | ||
| 15 | OIT3 | 1798 | 3.029 | 0.0839 | No | ||
| 16 | GRN | 1812 | 3.013 | 0.0916 | No | ||
| 17 | ANK3 | 2066 | 2.795 | 0.0877 | No | ||
| 18 | CTPS | 2231 | 2.682 | 0.0875 | No | ||
| 19 | TMLHE | 2260 | 2.667 | 0.0935 | No | ||
| 20 | LAMA2 | 2342 | 2.621 | 0.0970 | No | ||
| 21 | MAPK10 | 2538 | 2.517 | 0.0949 | No | ||
| 22 | SEMA6D | 2637 | 2.461 | 0.0972 | No | ||
| 23 | KLF4 | 2912 | 2.337 | 0.0910 | No | ||
| 24 | CITED4 | 2978 | 2.311 | 0.0944 | No | ||
| 25 | SERPING1 | 3230 | 2.210 | 0.0889 | No | ||
| 26 | NFATC4 | 3239 | 2.205 | 0.0946 | No | ||
| 27 | SDC1 | 3246 | 2.202 | 0.1004 | No | ||
| 28 | S100A3 | 3278 | 2.189 | 0.1049 | No | ||
| 29 | PHOX2B | 3643 | 2.046 | 0.0938 | No | ||
| 30 | SNCB | 3675 | 2.034 | 0.0980 | No | ||
| 31 | A2BP1 | 4149 | 1.881 | 0.0814 | No | ||
| 32 | TENC1 | 4264 | 1.841 | 0.0812 | No | ||
| 33 | COL25A1 | 4318 | 1.824 | 0.0838 | No | ||
| 34 | IRF4 | 4324 | 1.821 | 0.0886 | No | ||
| 35 | HFE2 | 4365 | 1.808 | 0.0917 | No | ||
| 36 | KCNK4 | 4376 | 1.805 | 0.0962 | No | ||
| 37 | POU4F1 | 4414 | 1.797 | 0.0994 | No | ||
| 38 | HSF4 | 4454 | 1.782 | 0.1025 | No | ||
| 39 | PLAGL1 | 4924 | 1.627 | 0.0854 | No | ||
| 40 | CACNA1D | 5025 | 1.597 | 0.0852 | No | ||
| 41 | TCEA2 | 5178 | 1.549 | 0.0825 | No | ||
| 42 | APBB1 | 5695 | 1.410 | 0.0627 | No | ||
| 43 | FOXA3 | 5763 | 1.392 | 0.0634 | No | ||
| 44 | PHF15 | 5799 | 1.382 | 0.0656 | No | ||
| 45 | SLC38A5 | 6039 | 1.326 | 0.0583 | No | ||
| 46 | PCOLCE | 6163 | 1.292 | 0.0562 | No | ||
| 47 | C1QTNF1 | 6264 | 1.269 | 0.0550 | No | ||
| 48 | TNFRSF9 | 6326 | 1.255 | 0.0557 | No | ||
| 49 | RRAS | 6393 | 1.241 | 0.0561 | No | ||
| 50 | LIF | 6518 | 1.214 | 0.0537 | No | ||
| 51 | PROS1 | 6646 | 1.184 | 0.0511 | No | ||
| 52 | PRKAR2B | 6710 | 1.168 | 0.0514 | No | ||
| 53 | PSD | 6808 | 1.147 | 0.0501 | No | ||
| 54 | LIN28 | 6829 | 1.139 | 0.0523 | No | ||
| 55 | ECEL1 | 6945 | 1.112 | 0.0501 | No | ||
| 56 | GPR153 | 7164 | 1.058 | 0.0430 | No | ||
| 57 | BDNF | 7264 | 1.037 | 0.0413 | No | ||
| 58 | FLT1 | 7430 | 0.998 | 0.0364 | No | ||
| 59 | COLQ | 7478 | 0.990 | 0.0370 | No | ||
| 60 | ENPP2 | 7713 | 0.937 | 0.0288 | No | ||
| 61 | CREM | 7942 | 0.886 | 0.0208 | No | ||
| 62 | MASP2 | 8104 | 0.845 | 0.0157 | No | ||
| 63 | PSMD3 | 8256 | 0.811 | 0.0110 | No | ||
| 64 | SCN3B | 8267 | 0.809 | 0.0127 | No | ||
| 65 | TNFSF11 | 8436 | 0.772 | 0.0071 | No | ||
| 66 | FOS | 8574 | 0.740 | 0.0029 | No | ||
| 67 | UNC13B | 8704 | 0.712 | -0.0011 | No | ||
| 68 | BET1 | 8952 | 0.649 | -0.0107 | No | ||
| 69 | SCRG1 | 9321 | 0.565 | -0.0260 | No | ||
| 70 | PLSCR1 | 9477 | 0.529 | -0.0317 | No | ||
| 71 | LEP | 9609 | 0.492 | -0.0364 | No | ||
| 72 | AMBN | 9760 | 0.451 | -0.0420 | No | ||
| 73 | HSPC171 | 9803 | 0.437 | -0.0427 | No | ||
| 74 | TRIM15 | 10130 | 0.346 | -0.0568 | No | ||
| 75 | BLR1 | 10209 | 0.324 | -0.0595 | No | ||
| 76 | BATF | 10285 | 0.305 | -0.0621 | No | ||
| 77 | KCNN3 | 10298 | 0.301 | -0.0618 | No | ||
| 78 | SRPX | 10346 | 0.288 | -0.0632 | No | ||
| 79 | APBA1 | 10390 | 0.274 | -0.0644 | No | ||
| 80 | DGKA | 10432 | 0.260 | -0.0656 | No | ||
| 81 | RAB11B | 10622 | 0.207 | -0.0737 | No | ||
| 82 | GLRA2 | 10759 | 0.167 | -0.0795 | No | ||
| 83 | PVALB | 11047 | 0.076 | -0.0924 | No | ||
| 84 | GMPPB | 11376 | -0.024 | -0.1074 | No | ||
| 85 | CXCL11 | 11672 | -0.121 | -0.1207 | No | ||
| 86 | CSF3 | 11775 | -0.149 | -0.1249 | No | ||
| 87 | RARA | 11911 | -0.188 | -0.1306 | No | ||
| 88 | CISH | 11959 | -0.200 | -0.1322 | No | ||
| 89 | BBS1 | 11994 | -0.209 | -0.1332 | No | ||
| 90 | CORT | 12105 | -0.247 | -0.1376 | No | ||
| 91 | TAL1 | 12350 | -0.318 | -0.1479 | No | ||
| 92 | ADAMTS4 | 12406 | -0.335 | -0.1496 | No | ||
| 93 | WNT3 | 12450 | -0.349 | -0.1506 | No | ||
| 94 | PPARGC1B | 12459 | -0.351 | -0.1500 | No | ||
| 95 | HOXA2 | 12841 | -0.462 | -0.1662 | No | ||
| 96 | FAP | 12903 | -0.478 | -0.1677 | No | ||
| 97 | PPARD | 12952 | -0.490 | -0.1686 | No | ||
| 98 | GSK3A | 12974 | -0.496 | -0.1682 | No | ||
| 99 | FGA | 12998 | -0.504 | -0.1678 | No | ||
| 100 | SLITRK4 | 13000 | -0.504 | -0.1665 | No | ||
| 101 | F9 | 13132 | -0.540 | -0.1710 | No | ||
| 102 | EGFL6 | 13173 | -0.553 | -0.1714 | No | ||
| 103 | OGG1 | 13330 | -0.597 | -0.1769 | No | ||
| 104 | PDLIM1 | 13401 | -0.622 | -0.1784 | No | ||
| 105 | TRIM46 | 13543 | -0.661 | -0.1831 | No | ||
| 106 | SMPD1 | 13655 | -0.684 | -0.1863 | No | ||
| 107 | PRDM1 | 13700 | -0.699 | -0.1864 | No | ||
| 108 | DLX4 | 13986 | -0.774 | -0.1974 | No | ||
| 109 | CYP26A1 | 14082 | -0.800 | -0.1995 | No | ||
| 110 | OSM | 14179 | -0.824 | -0.2017 | No | ||
| 111 | MAFF | 14202 | -0.827 | -0.2004 | No | ||
| 112 | LAPTM4B | 14237 | -0.836 | -0.1997 | No | ||
| 113 | LSAMP | 14395 | -0.876 | -0.2045 | No | ||
| 114 | TLX2 | 14397 | -0.877 | -0.2021 | No | ||
| 115 | NFIL3 | 14638 | -0.941 | -0.2106 | No | ||
| 116 | PCSK2 | 14686 | -0.954 | -0.2101 | No | ||
| 117 | MRPL24 | 14711 | -0.961 | -0.2086 | No | ||
| 118 | PROK1 | 14713 | -0.961 | -0.2060 | No | ||
| 119 | ADCK4 | 14842 | -0.996 | -0.2091 | No | ||
| 120 | SIGIRR | 14909 | -1.021 | -0.2094 | No | ||
| 121 | MECP2 | 15244 | -1.114 | -0.2217 | No | ||
| 122 | RFX4 | 15426 | -1.162 | -0.2268 | No | ||
| 123 | LAMA1 | 15675 | -1.227 | -0.2348 | No | ||
| 124 | ZNF235 | 15691 | -1.230 | -0.2321 | No | ||
| 125 | PHC1 | 15865 | -1.276 | -0.2366 | No | ||
| 126 | ADAMTS9 | 16238 | -1.373 | -0.2499 | No | ||
| 127 | CCL5 | 16676 | -1.488 | -0.2659 | No | ||
| 128 | ADAMTSL3 | 16943 | -1.567 | -0.2738 | No | ||
| 129 | MAP2K3 | 17062 | -1.606 | -0.2748 | No | ||
| 130 | PURA | 17225 | -1.659 | -0.2777 | No | ||
| 131 | DLL4 | 17365 | -1.707 | -0.2794 | No | ||
| 132 | BCL6 | 17488 | -1.746 | -0.2802 | No | ||
| 133 | GRB10 | 17515 | -1.755 | -0.2766 | No | ||
| 134 | KRTCAP2 | 17664 | -1.804 | -0.2785 | No | ||
| 135 | ITPKC | 17818 | -1.857 | -0.2804 | Yes | ||
| 136 | VIP | 17918 | -1.895 | -0.2797 | Yes | ||
| 137 | NTF5 | 17983 | -1.915 | -0.2774 | Yes | ||
| 138 | SOCS2 | 18133 | -1.970 | -0.2789 | Yes | ||
| 139 | ABCC5 | 18155 | -1.979 | -0.2744 | Yes | ||
| 140 | NRP1 | 18183 | -1.990 | -0.2702 | Yes | ||
| 141 | ICAM1 | 18458 | -2.101 | -0.2770 | Yes | ||
| 142 | CPNE1 | 18502 | -2.122 | -0.2731 | Yes | ||
| 143 | NDUFS2 | 18504 | -2.123 | -0.2674 | Yes | ||
| 144 | TLR7 | 18655 | -2.186 | -0.2682 | Yes | ||
| 145 | NR4A2 | 18880 | -2.284 | -0.2723 | Yes | ||
| 146 | VTN | 18889 | -2.287 | -0.2664 | Yes | ||
| 147 | STC1 | 19088 | -2.384 | -0.2689 | Yes | ||
| 148 | PMCH | 19206 | -2.450 | -0.2676 | Yes | ||
| 149 | CEBPG | 19368 | -2.531 | -0.2680 | Yes | ||
| 150 | ZYX | 19453 | -2.577 | -0.2648 | Yes | ||
| 151 | TSGA14 | 19615 | -2.666 | -0.2649 | Yes | ||
| 152 | SYNPO | 19682 | -2.699 | -0.2605 | Yes | ||
| 153 | XRCC1 | 19870 | -2.809 | -0.2614 | Yes | ||
| 154 | DTX2 | 20022 | -2.906 | -0.2603 | Yes | ||
| 155 | NT5C2 | 20036 | -2.918 | -0.2529 | Yes | ||
| 156 | ARF3 | 20104 | -2.968 | -0.2479 | Yes | ||
| 157 | TMOD3 | 20178 | -3.015 | -0.2429 | Yes | ||
| 158 | SFRS12 | 20190 | -3.022 | -0.2352 | Yes | ||
| 159 | NCALD | 20192 | -3.022 | -0.2269 | Yes | ||
| 160 | IRF2BP1 | 20284 | -3.087 | -0.2226 | Yes | ||
| 161 | CHN2 | 20349 | -3.135 | -0.2169 | Yes | ||
| 162 | SLC9A1 | 20386 | -3.160 | -0.2099 | Yes | ||
| 163 | EIF4E | 20471 | -3.243 | -0.2049 | Yes | ||
| 164 | AGRP | 20506 | -3.281 | -0.1974 | Yes | ||
| 165 | IRF1 | 20521 | -3.289 | -0.1891 | Yes | ||
| 166 | ISGF3G | 20598 | -3.358 | -0.1833 | Yes | ||
| 167 | NR4A3 | 20751 | -3.509 | -0.1807 | Yes | ||
| 168 | ATP5H | 20804 | -3.572 | -0.1733 | Yes | ||
| 169 | USP25 | 20827 | -3.597 | -0.1644 | Yes | ||
| 170 | NFIA | 20953 | -3.751 | -0.1598 | Yes | ||
| 171 | KCNJ8 | 20982 | -3.792 | -0.1507 | Yes | ||
| 172 | ASXL1 | 21029 | -3.860 | -0.1422 | Yes | ||
| 173 | HACE1 | 21300 | -4.361 | -0.1427 | Yes | ||
| 174 | ELL | 21351 | -4.484 | -0.1327 | Yes | ||
| 175 | OLFML3 | 21478 | -4.805 | -0.1253 | Yes | ||
| 176 | SEC24C | 21517 | -4.946 | -0.1134 | Yes | ||
| 177 | SYMPK | 21535 | -5.012 | -0.1005 | Yes | ||
| 178 | WASPIP | 21612 | -5.284 | -0.0894 | Yes | ||
| 179 | USP52 | 21664 | -5.490 | -0.0767 | Yes | ||
| 180 | AP2S1 | 21760 | -6.131 | -0.0643 | Yes | ||
| 181 | IRF2 | 21846 | -7.452 | -0.0477 | Yes | ||
| 182 | MAP4K4 | 21855 | -7.660 | -0.0270 | Yes | ||
| 183 | PIK4CB | 21919 | -11.376 | 0.0013 | Yes |