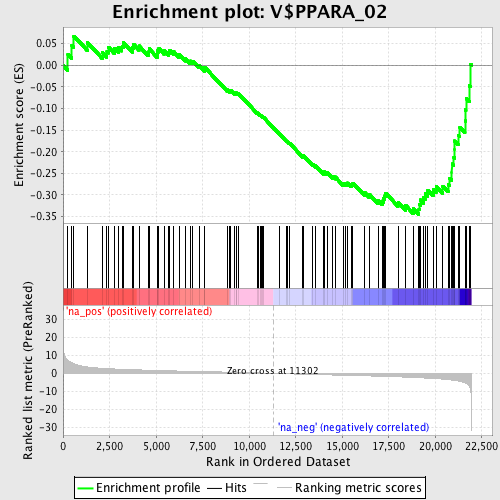
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set01_ATM_minus_versus_SCID |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_neg |
| GeneSet | V$PPARA_02 |
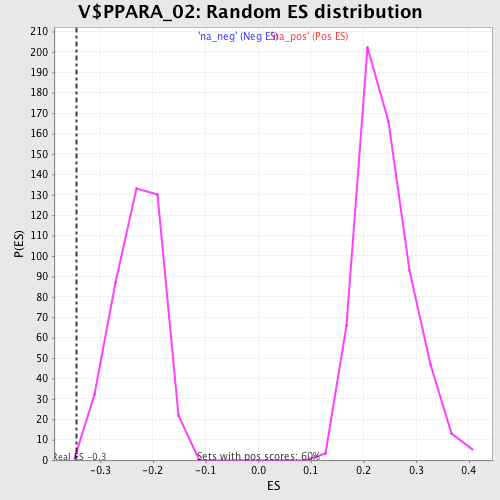
| Enrichment Score (ES) | -0.34507933 |
| Normalized Enrichment Score (NES) | -1.5111626 |
| Nominal p-value | 0.002469136 |
| FDR q-value | 0.37041283 |
| FWER p-Value | 0.88 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | RAD23B | 258 | 7.120 | 0.0250 | No | ||
| 2 | KPNB1 | 470 | 5.778 | 0.0452 | No | ||
| 3 | AIM1L | 576 | 5.219 | 0.0673 | No | ||
| 4 | PDGFB | 1313 | 3.537 | 0.0519 | No | ||
| 5 | SUPT16H | 2103 | 2.765 | 0.0301 | No | ||
| 6 | WNT4 | 2355 | 2.610 | 0.0321 | No | ||
| 7 | MAFB | 2443 | 2.564 | 0.0414 | No | ||
| 8 | KIF1C | 2753 | 2.408 | 0.0397 | No | ||
| 9 | PAK1 | 2989 | 2.306 | 0.0408 | No | ||
| 10 | ROM1 | 3164 | 2.234 | 0.0444 | No | ||
| 11 | KCNB1 | 3241 | 2.205 | 0.0523 | No | ||
| 12 | DHH | 3724 | 2.017 | 0.0407 | No | ||
| 13 | FTS | 3772 | 2.000 | 0.0489 | No | ||
| 14 | LAMA5 | 4079 | 1.899 | 0.0447 | No | ||
| 15 | RTN2 | 4581 | 1.739 | 0.0307 | No | ||
| 16 | FGF6 | 4614 | 1.726 | 0.0382 | No | ||
| 17 | HR | 5052 | 1.589 | 0.0264 | No | ||
| 18 | RIN1 | 5079 | 1.579 | 0.0334 | No | ||
| 19 | NEUROD2 | 5147 | 1.559 | 0.0384 | No | ||
| 20 | SEMA3A | 5424 | 1.484 | 0.0334 | No | ||
| 21 | TRIB1 | 5655 | 1.422 | 0.0302 | No | ||
| 22 | ASB7 | 5717 | 1.403 | 0.0347 | No | ||
| 23 | HOXA5 | 5927 | 1.350 | 0.0321 | No | ||
| 24 | NLGN3 | 6230 | 1.278 | 0.0249 | No | ||
| 25 | SERPINF2 | 6565 | 1.204 | 0.0158 | No | ||
| 26 | CDK5R2 | 6824 | 1.140 | 0.0099 | No | ||
| 27 | CACNA1S | 6955 | 1.109 | 0.0097 | No | ||
| 28 | PGF | 7302 | 1.028 | -0.0009 | No | ||
| 29 | GGN | 7592 | 0.965 | -0.0091 | No | ||
| 30 | FSCN3 | 7597 | 0.963 | -0.0043 | No | ||
| 31 | NEUROG1 | 8825 | 0.680 | -0.0569 | No | ||
| 32 | TRPV6 | 8943 | 0.652 | -0.0589 | No | ||
| 33 | TSNAXIP1 | 8993 | 0.640 | -0.0579 | No | ||
| 34 | TNXB | 9196 | 0.593 | -0.0640 | No | ||
| 35 | HOXA9 | 9294 | 0.572 | -0.0655 | No | ||
| 36 | CENTA2 | 9323 | 0.565 | -0.0639 | No | ||
| 37 | YARS | 9400 | 0.548 | -0.0645 | No | ||
| 38 | HMGN2 | 10426 | 0.262 | -0.1101 | No | ||
| 39 | NRGN | 10499 | 0.240 | -0.1121 | No | ||
| 40 | TITF1 | 10608 | 0.210 | -0.1160 | No | ||
| 41 | UCN2 | 10681 | 0.190 | -0.1183 | No | ||
| 42 | PSCA | 10736 | 0.177 | -0.1199 | No | ||
| 43 | PRX | 10781 | 0.160 | -0.1210 | No | ||
| 44 | HRH3 | 11624 | -0.104 | -0.1590 | No | ||
| 45 | PHKG2 | 11999 | -0.210 | -0.1751 | No | ||
| 46 | MNT | 12040 | -0.223 | -0.1757 | No | ||
| 47 | CYC1 | 12180 | -0.268 | -0.1807 | No | ||
| 48 | OTX1 | 12886 | -0.474 | -0.2105 | No | ||
| 49 | LMO1 | 12913 | -0.480 | -0.2092 | No | ||
| 50 | NKIRAS2 | 13419 | -0.627 | -0.2291 | No | ||
| 51 | CDH17 | 13568 | -0.667 | -0.2324 | No | ||
| 52 | EPHB1 | 14001 | -0.779 | -0.2482 | No | ||
| 53 | WDR20 | 14054 | -0.793 | -0.2465 | No | ||
| 54 | OSM | 14179 | -0.824 | -0.2479 | No | ||
| 55 | CREB3L2 | 14476 | -0.899 | -0.2568 | No | ||
| 56 | RGS8 | 14616 | -0.936 | -0.2583 | No | ||
| 57 | ERF | 15068 | -1.064 | -0.2734 | No | ||
| 58 | NXPH3 | 15182 | -1.097 | -0.2729 | No | ||
| 59 | PPP1R9B | 15294 | -1.128 | -0.2722 | No | ||
| 60 | ARHGAP26 | 15482 | -1.177 | -0.2747 | No | ||
| 61 | ESRRG | 15568 | -1.198 | -0.2724 | No | ||
| 62 | PKP3 | 16211 | -1.364 | -0.2947 | No | ||
| 63 | MSI1 | 16477 | -1.435 | -0.2994 | No | ||
| 64 | SLITRK5 | 16929 | -1.563 | -0.3120 | No | ||
| 65 | ATP5G2 | 17156 | -1.634 | -0.3139 | No | ||
| 66 | PURA | 17225 | -1.659 | -0.3084 | No | ||
| 67 | KRTAP6-3 | 17280 | -1.675 | -0.3022 | No | ||
| 68 | LOXL4 | 17325 | -1.691 | -0.2955 | No | ||
| 69 | ADD3 | 17996 | -1.918 | -0.3163 | No | ||
| 70 | VEGF | 18410 | -2.082 | -0.3244 | No | ||
| 71 | DDX17 | 18811 | -2.253 | -0.3311 | No | ||
| 72 | BAHD1 | 19118 | -2.400 | -0.3327 | Yes | ||
| 73 | CBX6 | 19150 | -2.417 | -0.3216 | Yes | ||
| 74 | RFX1 | 19204 | -2.448 | -0.3114 | Yes | ||
| 75 | EPN1 | 19369 | -2.531 | -0.3058 | Yes | ||
| 76 | SMARCA5 | 19457 | -2.580 | -0.2965 | Yes | ||
| 77 | PLAGL2 | 19597 | -2.657 | -0.2891 | Yes | ||
| 78 | PRKAG1 | 19884 | -2.818 | -0.2876 | Yes | ||
| 79 | POGZ | 20042 | -2.921 | -0.2797 | Yes | ||
| 80 | PERQ1 | 20403 | -3.173 | -0.2798 | Yes | ||
| 81 | DNAJC7 | 20690 | -3.440 | -0.2751 | Yes | ||
| 82 | POFUT1 | 20778 | -3.537 | -0.2608 | Yes | ||
| 83 | ZSWIM3 | 20893 | -3.676 | -0.2471 | Yes | ||
| 84 | CHD2 | 20895 | -3.680 | -0.2281 | Yes | ||
| 85 | RTN4 | 20996 | -3.813 | -0.2130 | Yes | ||
| 86 | HIP2 | 21042 | -3.880 | -0.1950 | Yes | ||
| 87 | MAP3K11 | 21046 | -3.888 | -0.1750 | Yes | ||
| 88 | ARF6 | 21229 | -4.197 | -0.1617 | Yes | ||
| 89 | DCTN1 | 21319 | -4.388 | -0.1431 | Yes | ||
| 90 | SCAMP3 | 21595 | -5.212 | -0.1287 | Yes | ||
| 91 | PHF12 | 21607 | -5.257 | -0.1021 | Yes | ||
| 92 | MAN2B1 | 21649 | -5.440 | -0.0759 | Yes | ||
| 93 | ABL1 | 21836 | -7.213 | -0.0471 | Yes | ||
| 94 | CTDSP1 | 21906 | -10.094 | 0.0019 | Yes |