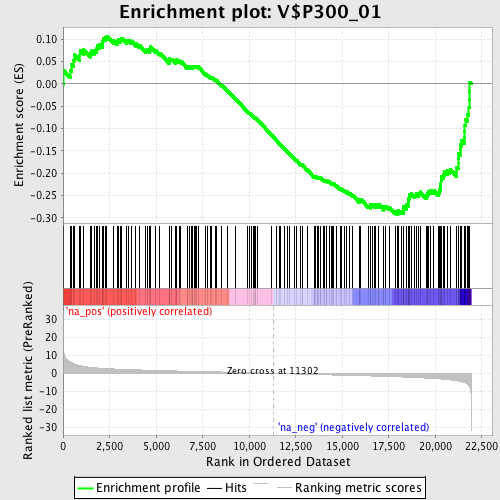
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set01_ATM_minus_versus_SCID |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_neg |
| GeneSet | V$P300_01 |
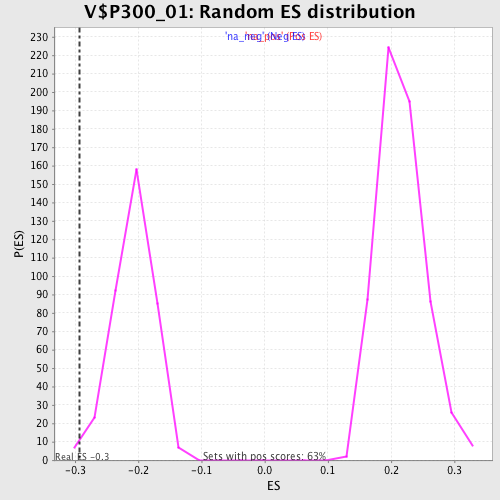
| Enrichment Score (ES) | -0.29257044 |
| Normalized Enrichment Score (NES) | -1.4065716 |
| Nominal p-value | 0.016129032 |
| FDR q-value | 0.42429748 |
| FWER p-Value | 0.992 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | SYNCRIP | 38 | 11.556 | 0.0298 | No | ||
| 2 | CANX | 388 | 6.232 | 0.0308 | No | ||
| 3 | ELAVL4 | 466 | 5.799 | 0.0431 | No | ||
| 4 | PPP1CB | 560 | 5.310 | 0.0533 | No | ||
| 5 | PSMD13 | 603 | 5.104 | 0.0653 | No | ||
| 6 | JARID2 | 891 | 4.340 | 0.0640 | No | ||
| 7 | ATP8A2 | 907 | 4.285 | 0.0750 | No | ||
| 8 | E2F3 | 1108 | 3.880 | 0.0764 | No | ||
| 9 | STAG2 | 1457 | 3.358 | 0.0695 | No | ||
| 10 | NLK | 1550 | 3.262 | 0.0742 | No | ||
| 11 | VAX1 | 1706 | 3.105 | 0.0756 | No | ||
| 12 | IKBKB | 1816 | 3.007 | 0.0788 | No | ||
| 13 | ELMO1 | 1838 | 2.992 | 0.0860 | No | ||
| 14 | DPP3 | 1958 | 2.878 | 0.0884 | No | ||
| 15 | EBAG9 | 2117 | 2.759 | 0.0886 | No | ||
| 16 | PPP3CB | 2125 | 2.754 | 0.0958 | No | ||
| 17 | PNOC | 2145 | 2.737 | 0.1024 | No | ||
| 18 | CAST | 2257 | 2.669 | 0.1046 | No | ||
| 19 | WNT4 | 2355 | 2.610 | 0.1073 | No | ||
| 20 | CHD4 | 2724 | 2.423 | 0.0970 | No | ||
| 21 | BCL11A | 2920 | 2.333 | 0.0944 | No | ||
| 22 | PPP1R1B | 2958 | 2.318 | 0.0990 | No | ||
| 23 | SP6 | 3091 | 2.262 | 0.0992 | No | ||
| 24 | HMGCS1 | 3145 | 2.242 | 0.1028 | No | ||
| 25 | CACNB2 | 3410 | 2.138 | 0.0966 | No | ||
| 26 | HOXC6 | 3502 | 2.101 | 0.0981 | No | ||
| 27 | GATA1 | 3662 | 2.039 | 0.0964 | No | ||
| 28 | GJB2 | 3911 | 1.955 | 0.0903 | No | ||
| 29 | ASCL1 | 4117 | 1.889 | 0.0861 | No | ||
| 30 | GRM7 | 4452 | 1.782 | 0.0756 | No | ||
| 31 | FGFR1 | 4520 | 1.764 | 0.0773 | No | ||
| 32 | LBX1 | 4663 | 1.710 | 0.0755 | No | ||
| 33 | ZIC4 | 4675 | 1.708 | 0.0796 | No | ||
| 34 | ADAM15 | 4685 | 1.706 | 0.0839 | No | ||
| 35 | HEBP1 | 4976 | 1.614 | 0.0750 | No | ||
| 36 | NR2F1 | 5195 | 1.544 | 0.0692 | No | ||
| 37 | TNFRSF12A | 5696 | 1.410 | 0.0500 | No | ||
| 38 | SUMO2 | 5716 | 1.403 | 0.0530 | No | ||
| 39 | ZCCHC5 | 5720 | 1.403 | 0.0567 | No | ||
| 40 | DPF3 | 5833 | 1.374 | 0.0553 | No | ||
| 41 | ZNF23 | 6033 | 1.327 | 0.0498 | No | ||
| 42 | GJB1 | 6071 | 1.319 | 0.0517 | No | ||
| 43 | PLP1 | 6098 | 1.313 | 0.0541 | No | ||
| 44 | NLGN3 | 6230 | 1.278 | 0.0515 | No | ||
| 45 | WNT7B | 6332 | 1.254 | 0.0503 | No | ||
| 46 | GSN | 6663 | 1.179 | 0.0384 | No | ||
| 47 | EPN3 | 6707 | 1.168 | 0.0396 | No | ||
| 48 | GDNF | 6792 | 1.149 | 0.0389 | No | ||
| 49 | MOAP1 | 6886 | 1.125 | 0.0377 | No | ||
| 50 | DMD | 6960 | 1.109 | 0.0373 | No | ||
| 51 | S100A16 | 6976 | 1.106 | 0.0397 | No | ||
| 52 | ZIC1 | 7036 | 1.090 | 0.0399 | No | ||
| 53 | HCN3 | 7116 | 1.070 | 0.0392 | No | ||
| 54 | SIX4 | 7188 | 1.054 | 0.0389 | No | ||
| 55 | CTBP2 | 7262 | 1.037 | 0.0383 | No | ||
| 56 | MYCL1 | 7663 | 0.947 | 0.0225 | No | ||
| 57 | POLD4 | 7773 | 0.926 | 0.0201 | No | ||
| 58 | KIF3B | 7924 | 0.890 | 0.0156 | No | ||
| 59 | NXPH4 | 7989 | 0.873 | 0.0150 | No | ||
| 60 | ARHGAP1 | 8167 | 0.831 | 0.0092 | No | ||
| 61 | WNT11 | 8239 | 0.814 | 0.0081 | No | ||
| 62 | INHA | 8524 | 0.755 | -0.0028 | No | ||
| 63 | NEUROG1 | 8825 | 0.680 | -0.0148 | No | ||
| 64 | STARD13 | 9248 | 0.582 | -0.0325 | No | ||
| 65 | DDIT4 | 9891 | 0.412 | -0.0609 | No | ||
| 66 | GPLD1 | 10024 | 0.376 | -0.0659 | No | ||
| 67 | SLC12A4 | 10138 | 0.343 | -0.0702 | No | ||
| 68 | TFEB | 10231 | 0.320 | -0.0735 | No | ||
| 69 | KCNN3 | 10298 | 0.301 | -0.0758 | No | ||
| 70 | ZHX2 | 10314 | 0.296 | -0.0756 | No | ||
| 71 | RHOV | 10349 | 0.287 | -0.0764 | No | ||
| 72 | DGKA | 10432 | 0.260 | -0.0795 | No | ||
| 73 | NKX6-2 | 11205 | 0.029 | -0.1148 | No | ||
| 74 | TRIM3 | 11439 | -0.047 | -0.1254 | No | ||
| 75 | TWIST1 | 11637 | -0.106 | -0.1342 | No | ||
| 76 | SEMA4C | 11661 | -0.115 | -0.1349 | No | ||
| 77 | RARA | 11911 | -0.188 | -0.1458 | No | ||
| 78 | VMD2L1 | 12044 | -0.223 | -0.1513 | No | ||
| 79 | RARG | 12185 | -0.271 | -0.1570 | No | ||
| 80 | WNT3 | 12450 | -0.349 | -0.1682 | No | ||
| 81 | FDPS | 12555 | -0.380 | -0.1719 | No | ||
| 82 | ACIN1 | 12777 | -0.444 | -0.1808 | No | ||
| 83 | HOXA2 | 12841 | -0.462 | -0.1825 | No | ||
| 84 | SREBF2 | 12843 | -0.462 | -0.1813 | No | ||
| 85 | FBXL19 | 13115 | -0.535 | -0.1922 | No | ||
| 86 | BHLHB3 | 13494 | -0.647 | -0.2078 | No | ||
| 87 | MTA2 | 13520 | -0.656 | -0.2072 | No | ||
| 88 | GFRA3 | 13586 | -0.671 | -0.2083 | No | ||
| 89 | BLCAP | 13659 | -0.687 | -0.2098 | No | ||
| 90 | TUSC3 | 13679 | -0.693 | -0.2088 | No | ||
| 91 | FZD5 | 13726 | -0.706 | -0.2089 | No | ||
| 92 | FURIN | 13813 | -0.726 | -0.2109 | No | ||
| 93 | HOXC4 | 13823 | -0.729 | -0.2093 | No | ||
| 94 | EPHB1 | 14001 | -0.779 | -0.2153 | No | ||
| 95 | AAMP | 14068 | -0.796 | -0.2162 | No | ||
| 96 | FSHB | 14147 | -0.816 | -0.2175 | No | ||
| 97 | WNT10A | 14165 | -0.821 | -0.2161 | No | ||
| 98 | EGR3 | 14289 | -0.850 | -0.2194 | No | ||
| 99 | NFKBIA | 14429 | -0.885 | -0.2234 | No | ||
| 100 | IRAK1 | 14459 | -0.896 | -0.2223 | No | ||
| 101 | SLITRK1 | 14536 | -0.916 | -0.2233 | No | ||
| 102 | NME1 | 14665 | -0.949 | -0.2266 | No | ||
| 103 | ZIC3 | 14895 | -1.014 | -0.2343 | No | ||
| 104 | EFNB3 | 14973 | -1.040 | -0.2350 | No | ||
| 105 | SLC9A6 | 15099 | -1.072 | -0.2378 | No | ||
| 106 | DMTF1 | 15245 | -1.114 | -0.2414 | No | ||
| 107 | SIRT3 | 15407 | -1.155 | -0.2457 | No | ||
| 108 | ESRRG | 15568 | -1.198 | -0.2498 | No | ||
| 109 | EMCN | 15908 | -1.286 | -0.2618 | No | ||
| 110 | PET112L | 15941 | -1.296 | -0.2597 | No | ||
| 111 | PRKAG2 | 15997 | -1.310 | -0.2587 | No | ||
| 112 | LDOC1 | 16415 | -1.420 | -0.2740 | No | ||
| 113 | MAP3K4 | 16508 | -1.443 | -0.2743 | No | ||
| 114 | GABRA1 | 16514 | -1.444 | -0.2706 | No | ||
| 115 | FBS1 | 16604 | -1.470 | -0.2706 | No | ||
| 116 | CKS1B | 16730 | -1.506 | -0.2723 | No | ||
| 117 | SMPD3 | 16794 | -1.523 | -0.2710 | No | ||
| 118 | SLITRK5 | 16929 | -1.563 | -0.2729 | No | ||
| 119 | KCNH3 | 16969 | -1.576 | -0.2704 | No | ||
| 120 | CSMD3 | 17238 | -1.661 | -0.2781 | No | ||
| 121 | SDF2 | 17240 | -1.661 | -0.2737 | No | ||
| 122 | SHC1 | 17343 | -1.699 | -0.2737 | No | ||
| 123 | PHC3 | 17522 | -1.758 | -0.2771 | No | ||
| 124 | LHX6 | 17837 | -1.865 | -0.2864 | No | ||
| 125 | LTBR | 17972 | -1.911 | -0.2874 | Yes | ||
| 126 | RGS14 | 17994 | -1.918 | -0.2831 | Yes | ||
| 127 | AKAP12 | 18173 | -1.985 | -0.2858 | Yes | ||
| 128 | CRIM1 | 18274 | -2.024 | -0.2849 | Yes | ||
| 129 | PIK3CD | 18281 | -2.027 | -0.2797 | Yes | ||
| 130 | BCL2L1 | 18302 | -2.040 | -0.2750 | Yes | ||
| 131 | SYN1 | 18436 | -2.092 | -0.2754 | Yes | ||
| 132 | JUNB | 18468 | -2.107 | -0.2711 | Yes | ||
| 133 | PTBP2 | 18539 | -2.135 | -0.2685 | Yes | ||
| 134 | RORC | 18540 | -2.135 | -0.2626 | Yes | ||
| 135 | GPC3 | 18559 | -2.141 | -0.2576 | Yes | ||
| 136 | DDAH2 | 18588 | -2.150 | -0.2530 | Yes | ||
| 137 | GRK5 | 18611 | -2.162 | -0.2481 | Yes | ||
| 138 | PUM2 | 18692 | -2.203 | -0.2458 | Yes | ||
| 139 | SUPT6H | 18904 | -2.296 | -0.2492 | Yes | ||
| 140 | PPP3CA | 18969 | -2.322 | -0.2458 | Yes | ||
| 141 | BAHD1 | 19118 | -2.400 | -0.2461 | Yes | ||
| 142 | NPR1 | 19177 | -2.432 | -0.2421 | Yes | ||
| 143 | FGF12 | 19506 | -2.610 | -0.2500 | Yes | ||
| 144 | EMP1 | 19577 | -2.647 | -0.2460 | Yes | ||
| 145 | RFX2 | 19634 | -2.672 | -0.2413 | Yes | ||
| 146 | MAP4K2 | 19737 | -2.734 | -0.2385 | Yes | ||
| 147 | KCNAB2 | 19895 | -2.825 | -0.2380 | Yes | ||
| 148 | RBMS2 | 20172 | -3.013 | -0.2425 | Yes | ||
| 149 | RERE | 20219 | -3.042 | -0.2363 | Yes | ||
| 150 | RASGRP2 | 20268 | -3.070 | -0.2301 | Yes | ||
| 151 | PCTK2 | 20299 | -3.099 | -0.2230 | Yes | ||
| 152 | ZFYVE26 | 20304 | -3.101 | -0.2148 | Yes | ||
| 153 | SERPINI1 | 20341 | -3.130 | -0.2079 | Yes | ||
| 154 | PDCD10 | 20445 | -3.216 | -0.2038 | Yes | ||
| 155 | PICALM | 20475 | -3.247 | -0.1963 | Yes | ||
| 156 | WDTC1 | 20643 | -3.392 | -0.1947 | Yes | ||
| 157 | ENSA | 20788 | -3.553 | -0.1916 | Yes | ||
| 158 | CENTD2 | 21152 | -4.063 | -0.1972 | Yes | ||
| 159 | DLX1 | 21159 | -4.071 | -0.1864 | Yes | ||
| 160 | ARF6 | 21229 | -4.197 | -0.1781 | Yes | ||
| 161 | CXXC5 | 21232 | -4.207 | -0.1667 | Yes | ||
| 162 | SRPK2 | 21253 | -4.263 | -0.1560 | Yes | ||
| 163 | PPP4C | 21328 | -4.405 | -0.1474 | Yes | ||
| 164 | FOXP1 | 21341 | -4.453 | -0.1358 | Yes | ||
| 165 | PRKACB | 21416 | -4.640 | -0.1265 | Yes | ||
| 166 | CAMK2D | 21559 | -5.093 | -0.1191 | Yes | ||
| 167 | ZBTB9 | 21575 | -5.151 | -0.1057 | Yes | ||
| 168 | PRKCBP1 | 21590 | -5.204 | -0.0922 | Yes | ||
| 169 | TNFRSF1A | 21616 | -5.292 | -0.0789 | Yes | ||
| 170 | DPYSL2 | 21727 | -5.920 | -0.0678 | Yes | ||
| 171 | NFATC1 | 21808 | -6.771 | -0.0530 | Yes | ||
| 172 | ERBB2IP | 21820 | -6.977 | -0.0344 | Yes | ||
| 173 | PML | 21829 | -7.144 | -0.0153 | Yes | ||
| 174 | SFRS14 | 21852 | -7.575 | 0.0044 | Yes |