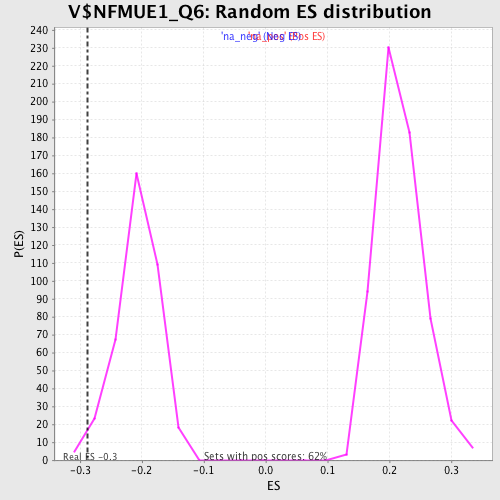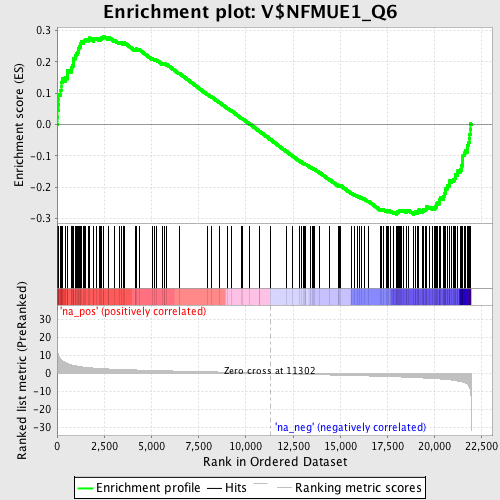
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set01_ATM_minus_versus_SCID |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_neg |
| GeneSet | V$NFMUE1_Q6 |
| Enrichment Score (ES) | -0.2872665 |
| Normalized Enrichment Score (NES) | -1.3880855 |
| Nominal p-value | 0.013089005 |
| FDR q-value | 0.3917958 |
| FWER p-Value | 0.997 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | SFRS10 | 19 | 13.401 | 0.0242 | No | ||
| 2 | EIF4G2 | 28 | 12.462 | 0.0472 | No | ||
| 3 | CCAR1 | 81 | 9.656 | 0.0629 | No | ||
| 4 | FKRP | 91 | 9.360 | 0.0800 | No | ||
| 5 | UBE2D3 | 95 | 9.312 | 0.0973 | No | ||
| 6 | NASP | 168 | 8.269 | 0.1095 | No | ||
| 7 | HNRPA1 | 209 | 7.587 | 0.1218 | No | ||
| 8 | EIF5 | 212 | 7.562 | 0.1359 | No | ||
| 9 | RIOK3 | 269 | 7.067 | 0.1466 | No | ||
| 10 | PRPS1 | 441 | 5.930 | 0.1498 | No | ||
| 11 | EIF3S10 | 536 | 5.426 | 0.1557 | No | ||
| 12 | BCLAF1 | 552 | 5.339 | 0.1650 | No | ||
| 13 | NCOR1 | 566 | 5.268 | 0.1743 | No | ||
| 14 | E2F6 | 739 | 4.692 | 0.1751 | No | ||
| 15 | STAG1 | 770 | 4.621 | 0.1824 | No | ||
| 16 | DHX15 | 802 | 4.544 | 0.1895 | No | ||
| 17 | WDR33 | 859 | 4.419 | 0.1952 | No | ||
| 18 | SRRM1 | 864 | 4.408 | 0.2033 | No | ||
| 19 | TCF4 | 887 | 4.351 | 0.2104 | No | ||
| 20 | BMI1 | 949 | 4.193 | 0.2155 | No | ||
| 21 | COL4A3BP | 981 | 4.127 | 0.2218 | No | ||
| 22 | REST | 1038 | 4.012 | 0.2267 | No | ||
| 23 | RAC1 | 1090 | 3.907 | 0.2317 | No | ||
| 24 | ATP6V0C | 1121 | 3.858 | 0.2375 | No | ||
| 25 | ARCN1 | 1141 | 3.819 | 0.2438 | No | ||
| 26 | EEF1A1 | 1172 | 3.763 | 0.2495 | No | ||
| 27 | YY1 | 1248 | 3.639 | 0.2528 | No | ||
| 28 | DAP3 | 1259 | 3.622 | 0.2592 | No | ||
| 29 | ZFX | 1275 | 3.602 | 0.2652 | No | ||
| 30 | ATP5F1 | 1382 | 3.446 | 0.2668 | No | ||
| 31 | POU2F1 | 1447 | 3.374 | 0.2702 | No | ||
| 32 | CUGBP1 | 1522 | 3.289 | 0.2729 | No | ||
| 33 | SFPQ | 1655 | 3.149 | 0.2728 | No | ||
| 34 | CPSF2 | 1696 | 3.111 | 0.2768 | No | ||
| 35 | CCNE1 | 1931 | 2.912 | 0.2715 | No | ||
| 36 | CLK1 | 1948 | 2.890 | 0.2762 | No | ||
| 37 | OSR1 | 2071 | 2.789 | 0.2758 | No | ||
| 38 | TNKS2 | 2253 | 2.672 | 0.2725 | No | ||
| 39 | ATF1 | 2314 | 2.637 | 0.2746 | No | ||
| 40 | MYNN | 2371 | 2.603 | 0.2770 | No | ||
| 41 | XPR1 | 2434 | 2.569 | 0.2789 | No | ||
| 42 | FBXL11 | 2459 | 2.558 | 0.2826 | No | ||
| 43 | JARID1C | 2720 | 2.424 | 0.2752 | No | ||
| 44 | VDAC2 | 2740 | 2.418 | 0.2789 | No | ||
| 45 | CLSTN1 | 3024 | 2.288 | 0.2701 | No | ||
| 46 | PCF11 | 3289 | 2.185 | 0.2621 | No | ||
| 47 | HNRPD | 3388 | 2.146 | 0.2616 | No | ||
| 48 | EP300 | 3501 | 2.101 | 0.2604 | No | ||
| 49 | PSMD8 | 3556 | 2.078 | 0.2618 | No | ||
| 50 | RAD23A | 4127 | 1.886 | 0.2392 | No | ||
| 51 | USF1 | 4175 | 1.871 | 0.2405 | No | ||
| 52 | BRD3 | 4189 | 1.866 | 0.2434 | No | ||
| 53 | PRPSAP1 | 4366 | 1.808 | 0.2387 | No | ||
| 54 | HMGB3 | 5053 | 1.589 | 0.2102 | No | ||
| 55 | NEUROD2 | 5147 | 1.559 | 0.2088 | No | ||
| 56 | PPP1R15B | 5272 | 1.524 | 0.2060 | No | ||
| 57 | SUV39H2 | 5592 | 1.435 | 0.1940 | No | ||
| 58 | ASB2 | 5672 | 1.415 | 0.1931 | No | ||
| 59 | APBB1 | 5695 | 1.410 | 0.1947 | No | ||
| 60 | PHF15 | 5799 | 1.382 | 0.1926 | No | ||
| 61 | CDH23 | 6468 | 1.225 | 0.1642 | No | ||
| 62 | RPL30 | 7955 | 0.882 | 0.0976 | No | ||
| 63 | ACTR8 | 8166 | 0.831 | 0.0895 | No | ||
| 64 | PCBP4 | 8607 | 0.733 | 0.0707 | No | ||
| 65 | ATF4 | 9033 | 0.631 | 0.0523 | No | ||
| 66 | MARK3 | 9246 | 0.583 | 0.0437 | No | ||
| 67 | THAP1 | 9786 | 0.442 | 0.0197 | No | ||
| 68 | ERH | 9802 | 0.437 | 0.0199 | No | ||
| 69 | SNAP25 | 10177 | 0.331 | 0.0033 | No | ||
| 70 | FBXO11 | 10735 | 0.177 | -0.0219 | No | ||
| 71 | NACA | 10740 | 0.176 | -0.0218 | No | ||
| 72 | CSAD | 11332 | -0.009 | -0.0489 | No | ||
| 73 | ZADH2 | 12155 | -0.261 | -0.0862 | No | ||
| 74 | MAEA | 12454 | -0.350 | -0.0992 | No | ||
| 75 | HOXA2 | 12841 | -0.462 | -0.1161 | No | ||
| 76 | WBSCR16 | 12927 | -0.485 | -0.1191 | No | ||
| 77 | UQCRH | 13079 | -0.527 | -0.1250 | No | ||
| 78 | NFYC | 13127 | -0.539 | -0.1262 | No | ||
| 79 | SNRPF | 13159 | -0.548 | -0.1266 | No | ||
| 80 | PRKCSH | 13182 | -0.555 | -0.1265 | No | ||
| 81 | NKIRAS2 | 13419 | -0.627 | -0.1362 | No | ||
| 82 | PSMB5 | 13523 | -0.657 | -0.1397 | No | ||
| 83 | CCND1 | 13583 | -0.671 | -0.1412 | No | ||
| 84 | PPRC1 | 13636 | -0.681 | -0.1423 | No | ||
| 85 | OTX2 | 13912 | -0.757 | -0.1535 | No | ||
| 86 | IRAK1 | 14459 | -0.896 | -0.1769 | No | ||
| 87 | PIGL | 14885 | -1.011 | -0.1945 | No | ||
| 88 | ZIC3 | 14895 | -1.014 | -0.1930 | No | ||
| 89 | MID1 | 14980 | -1.042 | -0.1949 | No | ||
| 90 | TMEM15 | 15041 | -1.057 | -0.1957 | No | ||
| 91 | POLK | 15603 | -1.208 | -0.2192 | No | ||
| 92 | RBM19 | 15738 | -1.241 | -0.2230 | No | ||
| 93 | ARF1 | 15903 | -1.285 | -0.2282 | No | ||
| 94 | PIGN | 16004 | -1.312 | -0.2303 | No | ||
| 95 | CSTF2T | 16125 | -1.340 | -0.2333 | No | ||
| 96 | NCDN | 16277 | -1.383 | -0.2376 | No | ||
| 97 | H1F0 | 16483 | -1.436 | -0.2444 | No | ||
| 98 | ATP5G2 | 17156 | -1.634 | -0.2722 | No | ||
| 99 | USP48 | 17178 | -1.641 | -0.2701 | No | ||
| 100 | NDST2 | 17303 | -1.683 | -0.2726 | No | ||
| 101 | SMYD5 | 17428 | -1.728 | -0.2751 | No | ||
| 102 | PHC3 | 17522 | -1.758 | -0.2760 | No | ||
| 103 | PIAS3 | 17585 | -1.780 | -0.2756 | No | ||
| 104 | GNPAT | 17678 | -1.810 | -0.2764 | No | ||
| 105 | PBX3 | 17849 | -1.869 | -0.2807 | No | ||
| 106 | MRPL38 | 17993 | -1.918 | -0.2837 | Yes | ||
| 107 | TOP3A | 17995 | -1.918 | -0.2801 | Yes | ||
| 108 | SNX13 | 18051 | -1.942 | -0.2790 | Yes | ||
| 109 | RBM3 | 18079 | -1.950 | -0.2766 | Yes | ||
| 110 | RPA2 | 18125 | -1.967 | -0.2750 | Yes | ||
| 111 | TNRC15 | 18179 | -1.987 | -0.2737 | Yes | ||
| 112 | ADK | 18271 | -2.024 | -0.2741 | Yes | ||
| 113 | UBE4B | 18346 | -2.057 | -0.2736 | Yes | ||
| 114 | CPNE1 | 18502 | -2.122 | -0.2768 | Yes | ||
| 115 | PTBP2 | 18539 | -2.135 | -0.2744 | Yes | ||
| 116 | VAMP2 | 18615 | -2.163 | -0.2738 | Yes | ||
| 117 | EPC1 | 18893 | -2.291 | -0.2823 | Yes | ||
| 118 | RALA | 18965 | -2.321 | -0.2812 | Yes | ||
| 119 | FASTK | 18971 | -2.324 | -0.2771 | Yes | ||
| 120 | BAHD1 | 19118 | -2.400 | -0.2793 | Yes | ||
| 121 | RPS27 | 19139 | -2.412 | -0.2757 | Yes | ||
| 122 | ZFP91 | 19174 | -2.430 | -0.2727 | Yes | ||
| 123 | SF1 | 19340 | -2.515 | -0.2756 | Yes | ||
| 124 | MAML1 | 19398 | -2.548 | -0.2734 | Yes | ||
| 125 | RFX3 | 19424 | -2.560 | -0.2697 | Yes | ||
| 126 | NSD1 | 19541 | -2.628 | -0.2702 | Yes | ||
| 127 | ZFP37 | 19590 | -2.653 | -0.2674 | Yes | ||
| 128 | NFYA | 19591 | -2.654 | -0.2624 | Yes | ||
| 129 | RPL35A | 19735 | -2.733 | -0.2639 | Yes | ||
| 130 | RNF26 | 19876 | -2.812 | -0.2650 | Yes | ||
| 131 | APBB3 | 19995 | -2.892 | -0.2650 | Yes | ||
| 132 | RAB22A | 20059 | -2.931 | -0.2624 | Yes | ||
| 133 | STRN4 | 20086 | -2.958 | -0.2581 | Yes | ||
| 134 | ARNT | 20092 | -2.961 | -0.2528 | Yes | ||
| 135 | PPP1CC | 20164 | -3.005 | -0.2504 | Yes | ||
| 136 | PABPN1 | 20239 | -3.051 | -0.2481 | Yes | ||
| 137 | CBX5 | 20242 | -3.054 | -0.2425 | Yes | ||
| 138 | ASH1L | 20267 | -3.070 | -0.2378 | Yes | ||
| 139 | PPP1R12B | 20296 | -3.097 | -0.2333 | Yes | ||
| 140 | CD3E | 20464 | -3.238 | -0.2349 | Yes | ||
| 141 | PICALM | 20475 | -3.247 | -0.2293 | Yes | ||
| 142 | RIF1 | 20509 | -3.284 | -0.2247 | Yes | ||
| 143 | SLC35A4 | 20545 | -3.304 | -0.2201 | Yes | ||
| 144 | DGCR2 | 20567 | -3.325 | -0.2148 | Yes | ||
| 145 | E4F1 | 20574 | -3.335 | -0.2089 | Yes | ||
| 146 | WASL | 20582 | -3.340 | -0.2029 | Yes | ||
| 147 | RBM12 | 20662 | -3.415 | -0.2002 | Yes | ||
| 148 | DNAJC7 | 20690 | -3.440 | -0.1950 | Yes | ||
| 149 | PHF8 | 20770 | -3.528 | -0.1920 | Yes | ||
| 150 | ENSA | 20788 | -3.553 | -0.1861 | Yes | ||
| 151 | ATP5H | 20804 | -3.572 | -0.1801 | Yes | ||
| 152 | CHD2 | 20895 | -3.680 | -0.1773 | Yes | ||
| 153 | CD4 | 20985 | -3.795 | -0.1743 | Yes | ||
| 154 | DZIP1 | 21083 | -3.943 | -0.1714 | Yes | ||
| 155 | DEAF1 | 21105 | -3.978 | -0.1649 | Yes | ||
| 156 | DDX6 | 21125 | -4.012 | -0.1583 | Yes | ||
| 157 | ODF2 | 21216 | -4.174 | -0.1546 | Yes | ||
| 158 | RAB1A | 21240 | -4.235 | -0.1477 | Yes | ||
| 159 | ZZEF1 | 21354 | -4.495 | -0.1445 | Yes | ||
| 160 | SMCR8 | 21427 | -4.681 | -0.1390 | Yes | ||
| 161 | UTX | 21431 | -4.694 | -0.1304 | Yes | ||
| 162 | OLFML3 | 21478 | -4.805 | -0.1235 | Yes | ||
| 163 | UBE2J2 | 21484 | -4.825 | -0.1147 | Yes | ||
| 164 | AP3D1 | 21496 | -4.863 | -0.1061 | Yes | ||
| 165 | RBM5 | 21505 | -4.898 | -0.0973 | Yes | ||
| 166 | CNOT3 | 21570 | -5.136 | -0.0906 | Yes | ||
| 167 | RBM4 | 21617 | -5.297 | -0.0828 | Yes | ||
| 168 | TSC1 | 21728 | -5.921 | -0.0768 | Yes | ||
| 169 | TIA1 | 21757 | -6.125 | -0.0666 | Yes | ||
| 170 | GBF1 | 21781 | -6.360 | -0.0557 | Yes | ||
| 171 | PPARBP | 21840 | -7.274 | -0.0448 | Yes | ||
| 172 | BCL11B | 21858 | -7.702 | -0.0311 | Yes | ||
| 173 | EXT1 | 21887 | -8.877 | -0.0158 | Yes | ||
| 174 | RPAP1 | 21901 | -9.874 | 0.0021 | Yes |