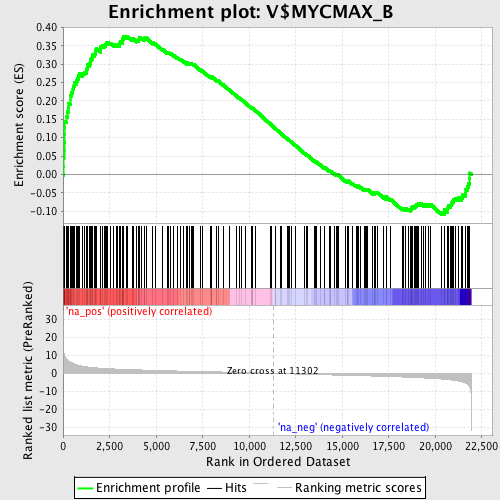
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set01_ATM_minus_versus_SCID |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_pos |
| GeneSet | V$MYCMAX_B |
| Enrichment Score (ES) | 0.3761045 |
| Normalized Enrichment Score (NES) | 1.7593864 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.04201005 |
| FWER p-Value | 0.243 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | NCL | 36 | 11.624 | 0.0218 | Yes | ||
| 2 | ABCE1 | 41 | 11.383 | 0.0445 | Yes | ||
| 3 | YTHDF2 | 48 | 11.012 | 0.0664 | Yes | ||
| 4 | SNX5 | 51 | 10.860 | 0.0882 | Yes | ||
| 5 | VAPA | 71 | 10.075 | 0.1076 | Yes | ||
| 6 | ETF1 | 77 | 9.911 | 0.1273 | Yes | ||
| 7 | UBE2D3 | 95 | 9.312 | 0.1453 | Yes | ||
| 8 | PABPC1 | 176 | 8.119 | 0.1580 | Yes | ||
| 9 | XRN2 | 224 | 7.473 | 0.1709 | Yes | ||
| 10 | RBBP7 | 304 | 6.782 | 0.1809 | Yes | ||
| 11 | SLC25A5 | 311 | 6.743 | 0.1942 | Yes | ||
| 12 | NPM1 | 398 | 6.184 | 0.2027 | Yes | ||
| 13 | CCT5 | 400 | 6.178 | 0.2151 | Yes | ||
| 14 | KPNB1 | 470 | 5.778 | 0.2236 | Yes | ||
| 15 | HNRPDL | 529 | 5.454 | 0.2319 | Yes | ||
| 16 | RPS6KA3 | 571 | 5.239 | 0.2406 | Yes | ||
| 17 | IVNS1ABP | 594 | 5.153 | 0.2499 | Yes | ||
| 18 | AK3 | 698 | 4.811 | 0.2549 | Yes | ||
| 19 | PGRMC2 | 771 | 4.620 | 0.2609 | Yes | ||
| 20 | ZNRF2 | 808 | 4.536 | 0.2684 | Yes | ||
| 21 | HNRPR | 858 | 4.419 | 0.2750 | Yes | ||
| 22 | ENPP6 | 1041 | 4.004 | 0.2747 | Yes | ||
| 23 | IPO7 | 1127 | 3.845 | 0.2785 | Yes | ||
| 24 | NELF | 1243 | 3.647 | 0.2806 | Yes | ||
| 25 | TGIF | 1266 | 3.612 | 0.2869 | Yes | ||
| 26 | PDGFB | 1313 | 3.537 | 0.2919 | Yes | ||
| 27 | SLC1A1 | 1317 | 3.530 | 0.2988 | Yes | ||
| 28 | CRK | 1421 | 3.408 | 0.3010 | Yes | ||
| 29 | STAG2 | 1457 | 3.358 | 0.3061 | Yes | ||
| 30 | PTMA | 1471 | 3.346 | 0.3123 | Yes | ||
| 31 | SEC61A1 | 1521 | 3.290 | 0.3166 | Yes | ||
| 32 | UBE2S | 1587 | 3.216 | 0.3201 | Yes | ||
| 33 | BZW2 | 1589 | 3.215 | 0.3266 | Yes | ||
| 34 | VAX1 | 1706 | 3.105 | 0.3275 | Yes | ||
| 35 | HMGA1 | 1732 | 3.086 | 0.3325 | Yes | ||
| 36 | NSF | 1740 | 3.077 | 0.3384 | Yes | ||
| 37 | DLGAP4 | 1781 | 3.046 | 0.3427 | Yes | ||
| 38 | SOX4 | 1998 | 2.850 | 0.3385 | Yes | ||
| 39 | EXOSC5 | 1999 | 2.849 | 0.3443 | Yes | ||
| 40 | CEBPA | 2031 | 2.828 | 0.3485 | Yes | ||
| 41 | EBAG9 | 2117 | 2.759 | 0.3502 | Yes | ||
| 42 | ALS2CR13 | 2234 | 2.681 | 0.3503 | Yes | ||
| 43 | HTF9C | 2271 | 2.662 | 0.3540 | Yes | ||
| 44 | ASCL2 | 2305 | 2.639 | 0.3578 | Yes | ||
| 45 | PAIP2 | 2387 | 2.594 | 0.3593 | Yes | ||
| 46 | PCTK1 | 2545 | 2.513 | 0.3571 | Yes | ||
| 47 | SDK1 | 2721 | 2.424 | 0.3539 | Yes | ||
| 48 | KIAA0427 | 2875 | 2.355 | 0.3516 | Yes | ||
| 49 | FCHSD2 | 2947 | 2.324 | 0.3531 | Yes | ||
| 50 | GTF2A1 | 3046 | 2.279 | 0.3531 | Yes | ||
| 51 | NR2E1 | 3053 | 2.277 | 0.3575 | Yes | ||
| 52 | OSBPL10 | 3058 | 2.275 | 0.3619 | Yes | ||
| 53 | ZF | 3177 | 2.229 | 0.3609 | Yes | ||
| 54 | INSM1 | 3181 | 2.227 | 0.3653 | Yes | ||
| 55 | EIF3S9 | 3206 | 2.218 | 0.3686 | Yes | ||
| 56 | RANBP1 | 3228 | 2.211 | 0.3721 | Yes | ||
| 57 | NFATC4 | 3239 | 2.205 | 0.3761 | Yes | ||
| 58 | HNRPD | 3388 | 2.146 | 0.3736 | No | ||
| 59 | SERPINF1 | 3464 | 2.120 | 0.3744 | No | ||
| 60 | SYT7 | 3706 | 2.024 | 0.3674 | No | ||
| 61 | CXCL14 | 3759 | 2.005 | 0.3691 | No | ||
| 62 | PHF7 | 3957 | 1.943 | 0.3639 | No | ||
| 63 | NEUROG3 | 4041 | 1.914 | 0.3640 | No | ||
| 64 | MMP11 | 4049 | 1.910 | 0.3675 | No | ||
| 65 | DAZL | 4088 | 1.898 | 0.3696 | No | ||
| 66 | ASCL1 | 4117 | 1.889 | 0.3721 | No | ||
| 67 | TAF11 | 4227 | 1.855 | 0.3708 | No | ||
| 68 | ACY1 | 4364 | 1.809 | 0.3682 | No | ||
| 69 | NKX2-8 | 4381 | 1.804 | 0.3711 | No | ||
| 70 | HSF4 | 4454 | 1.782 | 0.3714 | No | ||
| 71 | G6PC3 | 4821 | 1.662 | 0.3579 | No | ||
| 72 | RNF125 | 4954 | 1.622 | 0.3551 | No | ||
| 73 | SALL1 | 5345 | 1.505 | 0.3402 | No | ||
| 74 | ZDHHC5 | 5595 | 1.435 | 0.3317 | No | ||
| 75 | TRIB1 | 5655 | 1.422 | 0.3318 | No | ||
| 76 | WEE1 | 5782 | 1.386 | 0.3288 | No | ||
| 77 | RCL1 | 5955 | 1.343 | 0.3236 | No | ||
| 78 | JMJD1A | 6161 | 1.293 | 0.3168 | No | ||
| 79 | GRIN2B | 6287 | 1.264 | 0.3136 | No | ||
| 80 | ISL1 | 6476 | 1.223 | 0.3074 | No | ||
| 81 | PLCD1 | 6649 | 1.183 | 0.3019 | No | ||
| 82 | CPNE5 | 6658 | 1.180 | 0.3039 | No | ||
| 83 | BCKDHA | 6786 | 1.150 | 0.3004 | No | ||
| 84 | LRFN5 | 6816 | 1.144 | 0.3014 | No | ||
| 85 | SOX12 | 6877 | 1.128 | 0.3009 | No | ||
| 86 | HOXB9 | 6942 | 1.112 | 0.3002 | No | ||
| 87 | H1FX | 7009 | 1.097 | 0.2993 | No | ||
| 88 | EIF5A | 7362 | 1.015 | 0.2852 | No | ||
| 89 | KLF13 | 7470 | 0.991 | 0.2823 | No | ||
| 90 | RNF146 | 7899 | 0.895 | 0.2644 | No | ||
| 91 | NDUFS1 | 7965 | 0.880 | 0.2632 | No | ||
| 92 | DBP | 7974 | 0.877 | 0.2646 | No | ||
| 93 | RPS19 | 7991 | 0.872 | 0.2656 | No | ||
| 94 | EPHA2 | 8247 | 0.813 | 0.2555 | No | ||
| 95 | MICAL2 | 8265 | 0.810 | 0.2564 | No | ||
| 96 | XYLT2 | 8329 | 0.798 | 0.2551 | No | ||
| 97 | PCBP4 | 8607 | 0.733 | 0.2438 | No | ||
| 98 | PER1 | 8937 | 0.652 | 0.2300 | No | ||
| 99 | HOXA9 | 9294 | 0.572 | 0.2148 | No | ||
| 100 | TGIF2 | 9467 | 0.533 | 0.2080 | No | ||
| 101 | HOXC5 | 9580 | 0.499 | 0.2038 | No | ||
| 102 | NR1D1 | 9787 | 0.441 | 0.1953 | No | ||
| 103 | ZFP1 | 10134 | 0.345 | 0.1800 | No | ||
| 104 | WNT1 | 10135 | 0.345 | 0.1807 | No | ||
| 105 | RTBDN | 10150 | 0.338 | 0.1808 | No | ||
| 106 | SNAP25 | 10177 | 0.331 | 0.1802 | No | ||
| 107 | SIX1 | 10334 | 0.291 | 0.1737 | No | ||
| 108 | HPCAL4 | 11147 | 0.046 | 0.1364 | No | ||
| 109 | STMN1 | 11211 | 0.027 | 0.1336 | No | ||
| 110 | ALOXE3 | 11413 | -0.036 | 0.1244 | No | ||
| 111 | ID1 | 11419 | -0.040 | 0.1243 | No | ||
| 112 | CRABP2 | 11704 | -0.128 | 0.1115 | No | ||
| 113 | PTF1A | 11724 | -0.135 | 0.1109 | No | ||
| 114 | PITX3 | 12034 | -0.221 | 0.0971 | No | ||
| 115 | JUB | 12106 | -0.247 | 0.0943 | No | ||
| 116 | CALM2 | 12170 | -0.266 | 0.0920 | No | ||
| 117 | FOXA2 | 12246 | -0.285 | 0.0891 | No | ||
| 118 | PPARGC1B | 12459 | -0.351 | 0.0801 | No | ||
| 119 | NTN2L | 12966 | -0.493 | 0.0578 | No | ||
| 120 | SOCS3 | 13073 | -0.525 | 0.0540 | No | ||
| 121 | ZFP36L1 | 13142 | -0.543 | 0.0520 | No | ||
| 122 | BHLHB3 | 13494 | -0.647 | 0.0371 | No | ||
| 123 | CCND1 | 13583 | -0.671 | 0.0344 | No | ||
| 124 | PFN1 | 13606 | -0.674 | 0.0348 | No | ||
| 125 | HOXC4 | 13823 | -0.729 | 0.0263 | No | ||
| 126 | ELAVL3 | 14026 | -0.786 | 0.0186 | No | ||
| 127 | TCOF1 | 14050 | -0.792 | 0.0191 | No | ||
| 128 | EGR3 | 14289 | -0.850 | 0.0099 | No | ||
| 129 | SPG21 | 14353 | -0.867 | 0.0088 | No | ||
| 130 | PTK7 | 14392 | -0.875 | 0.0088 | No | ||
| 131 | CATSPER2 | 14559 | -0.920 | 0.0030 | No | ||
| 132 | SIX5 | 14712 | -0.961 | -0.0020 | No | ||
| 133 | ALDH1A2 | 14751 | -0.972 | -0.0018 | No | ||
| 134 | ILF3 | 14779 | -0.980 | -0.0011 | No | ||
| 135 | BAP1 | 15187 | -1.097 | -0.0176 | No | ||
| 136 | SCML2 | 15266 | -1.120 | -0.0189 | No | ||
| 137 | PPP1R9B | 15294 | -1.128 | -0.0179 | No | ||
| 138 | HPCA | 15332 | -1.136 | -0.0173 | No | ||
| 139 | NEUROD1 | 15550 | -1.194 | -0.0249 | No | ||
| 140 | SCRT2 | 15744 | -1.242 | -0.0313 | No | ||
| 141 | EEF1B2 | 15833 | -1.269 | -0.0327 | No | ||
| 142 | CRMP1 | 15852 | -1.274 | -0.0310 | No | ||
| 143 | PRKAG2 | 15997 | -1.310 | -0.0350 | No | ||
| 144 | TRIM8 | 16203 | -1.361 | -0.0417 | No | ||
| 145 | UBXD3 | 16240 | -1.373 | -0.0406 | No | ||
| 146 | DAZAP1 | 16317 | -1.395 | -0.0412 | No | ||
| 147 | HN1 | 16357 | -1.406 | -0.0402 | No | ||
| 148 | RKHD3 | 16645 | -1.480 | -0.0504 | No | ||
| 149 | LRP8 | 16728 | -1.505 | -0.0512 | No | ||
| 150 | ANKMY2 | 16744 | -1.510 | -0.0488 | No | ||
| 151 | SYT12 | 16801 | -1.524 | -0.0483 | No | ||
| 152 | EPS8 | 16866 | -1.545 | -0.0481 | No | ||
| 153 | GIT1 | 17216 | -1.655 | -0.0608 | No | ||
| 154 | TRPC4AP | 17353 | -1.703 | -0.0637 | No | ||
| 155 | DLL4 | 17365 | -1.707 | -0.0607 | No | ||
| 156 | SDHB | 17604 | -1.785 | -0.0681 | No | ||
| 157 | PLXNC1 | 18210 | -2.000 | -0.0919 | No | ||
| 158 | APLN | 18314 | -2.044 | -0.0925 | No | ||
| 159 | VEGF | 18410 | -2.082 | -0.0927 | No | ||
| 160 | RORC | 18540 | -2.135 | -0.0943 | No | ||
| 161 | RAPGEF6 | 18652 | -2.185 | -0.0950 | No | ||
| 162 | ABTB2 | 18693 | -2.203 | -0.0924 | No | ||
| 163 | ELOVL6 | 18737 | -2.219 | -0.0899 | No | ||
| 164 | FGF14 | 18792 | -2.246 | -0.0879 | No | ||
| 165 | EWSR1 | 18870 | -2.279 | -0.0868 | No | ||
| 166 | PTOV1 | 18927 | -2.304 | -0.0847 | No | ||
| 167 | GFI1 | 18975 | -2.325 | -0.0822 | No | ||
| 168 | RAI14 | 19036 | -2.357 | -0.0802 | No | ||
| 169 | IRX2 | 19087 | -2.384 | -0.0777 | No | ||
| 170 | SPAG9 | 19235 | -2.466 | -0.0795 | No | ||
| 171 | ARL3 | 19383 | -2.538 | -0.0812 | No | ||
| 172 | SRF | 19495 | -2.604 | -0.0810 | No | ||
| 173 | SMAD7 | 19639 | -2.675 | -0.0822 | No | ||
| 174 | MYOHD1 | 19747 | -2.738 | -0.0816 | No | ||
| 175 | SNRP70 | 20333 | -3.122 | -0.1022 | No | ||
| 176 | PICALM | 20475 | -3.247 | -0.1022 | No | ||
| 177 | DNAJC11 | 20482 | -3.255 | -0.0959 | No | ||
| 178 | KCNH2 | 20668 | -3.418 | -0.0975 | No | ||
| 179 | HS3ST3B1 | 20678 | -3.426 | -0.0910 | No | ||
| 180 | PIP5K2B | 20683 | -3.430 | -0.0843 | No | ||
| 181 | CALM3 | 20830 | -3.605 | -0.0837 | No | ||
| 182 | NUTF2 | 20850 | -3.630 | -0.0773 | No | ||
| 183 | BRUNOL4 | 20915 | -3.703 | -0.0728 | No | ||
| 184 | PES1 | 20968 | -3.769 | -0.0676 | No | ||
| 185 | SPOP | 21064 | -3.916 | -0.0641 | No | ||
| 186 | ARF6 | 21229 | -4.197 | -0.0631 | No | ||
| 187 | ZDHHC14 | 21422 | -4.658 | -0.0626 | No | ||
| 188 | ANAPC10 | 21480 | -4.809 | -0.0555 | No | ||
| 189 | UNC84B | 21599 | -5.236 | -0.0504 | No | ||
| 190 | SEMA7A | 21638 | -5.380 | -0.0413 | No | ||
| 191 | KIT | 21712 | -5.754 | -0.0331 | No | ||
| 192 | TNPO2 | 21787 | -6.441 | -0.0235 | No | ||
| 193 | SFRS14 | 21852 | -7.575 | -0.0112 | No | ||
| 194 | BCL11B | 21858 | -7.702 | 0.0041 | No |