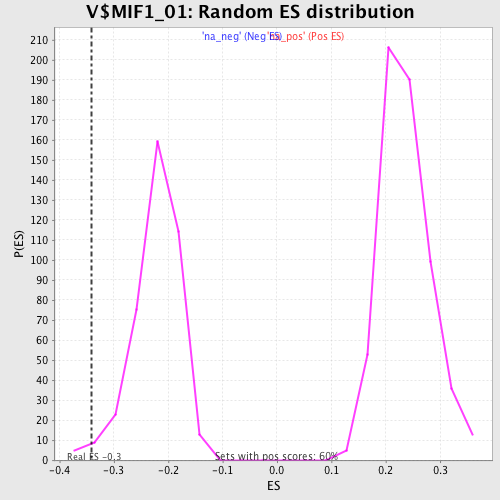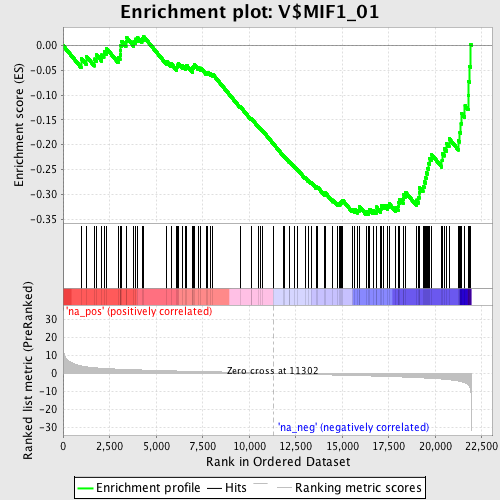
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set01_ATM_minus_versus_SCID |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_neg |
| GeneSet | V$MIF1_01 |
| Enrichment Score (ES) | -0.3408579 |
| Normalized Enrichment Score (NES) | -1.5346537 |
| Nominal p-value | 0.012562814 |
| FDR q-value | 0.493898 |
| FWER p-Value | 0.806 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | HM13 | 972 | 4.137 | -0.0256 | No | ||
| 2 | NELF | 1243 | 3.647 | -0.0214 | No | ||
| 3 | CX3CL1 | 1680 | 3.131 | -0.0271 | No | ||
| 4 | CHCHD4 | 1799 | 3.028 | -0.0187 | No | ||
| 5 | TPCN1 | 2082 | 2.780 | -0.0189 | No | ||
| 6 | WWP1 | 2197 | 2.700 | -0.0118 | No | ||
| 7 | SMAD5 | 2331 | 2.629 | -0.0059 | No | ||
| 8 | TLX3 | 2959 | 2.318 | -0.0240 | No | ||
| 9 | RASGEF1A | 3061 | 2.274 | -0.0183 | No | ||
| 10 | TBPL1 | 3086 | 2.263 | -0.0091 | No | ||
| 11 | MAGED1 | 3103 | 2.258 | 0.0005 | No | ||
| 12 | HMGCS1 | 3145 | 2.242 | 0.0088 | No | ||
| 13 | ELAVL2 | 3380 | 2.149 | 0.0079 | No | ||
| 14 | TTC8 | 3395 | 2.142 | 0.0171 | No | ||
| 15 | HAT1 | 3804 | 1.990 | 0.0074 | No | ||
| 16 | SOX5 | 3874 | 1.966 | 0.0133 | No | ||
| 17 | KIF17 | 4008 | 1.925 | 0.0159 | No | ||
| 18 | SOX3 | 4239 | 1.852 | 0.0139 | No | ||
| 19 | NAT5 | 4309 | 1.827 | 0.0190 | No | ||
| 20 | TBC1D16 | 5551 | 1.446 | -0.0312 | No | ||
| 21 | PHF15 | 5799 | 1.382 | -0.0362 | No | ||
| 22 | FILIP1 | 6097 | 1.313 | -0.0438 | No | ||
| 23 | BBS2 | 6125 | 1.304 | -0.0391 | No | ||
| 24 | CCND2 | 6194 | 1.286 | -0.0364 | No | ||
| 25 | MYCBP | 6407 | 1.239 | -0.0404 | No | ||
| 26 | SPRY2 | 6601 | 1.196 | -0.0438 | No | ||
| 27 | PARK2 | 6628 | 1.190 | -0.0396 | No | ||
| 28 | DMD | 6960 | 1.109 | -0.0497 | No | ||
| 29 | GRWD1 | 6962 | 1.108 | -0.0447 | No | ||
| 30 | SDCBP2 | 7012 | 1.096 | -0.0419 | No | ||
| 31 | DNAH9 | 7035 | 1.090 | -0.0379 | No | ||
| 32 | BDNF | 7264 | 1.037 | -0.0437 | No | ||
| 33 | MDH1B | 7400 | 1.005 | -0.0453 | No | ||
| 34 | PLA2G12A | 7684 | 0.942 | -0.0539 | No | ||
| 35 | POLD4 | 7773 | 0.926 | -0.0537 | No | ||
| 36 | KIF3B | 7924 | 0.890 | -0.0565 | No | ||
| 37 | CRLF1 | 8038 | 0.861 | -0.0578 | No | ||
| 38 | DHRS3 | 9518 | 0.517 | -0.1232 | No | ||
| 39 | PTCH2 | 10097 | 0.355 | -0.1480 | No | ||
| 40 | GPRC5D | 10127 | 0.346 | -0.1478 | No | ||
| 41 | NRGN | 10499 | 0.240 | -0.1637 | No | ||
| 42 | GLRA3 | 10586 | 0.216 | -0.1666 | No | ||
| 43 | GRIK5 | 10737 | 0.176 | -0.1727 | No | ||
| 44 | SALL2 | 11313 | -0.003 | -0.1990 | No | ||
| 45 | TEKT2 | 11824 | -0.163 | -0.2216 | No | ||
| 46 | BMP6 | 11908 | -0.187 | -0.2246 | No | ||
| 47 | TTC16 | 12152 | -0.261 | -0.2345 | No | ||
| 48 | FIBCD1 | 12190 | -0.272 | -0.2350 | No | ||
| 49 | STMN4 | 12443 | -0.347 | -0.2449 | No | ||
| 50 | THNSL1 | 12586 | -0.390 | -0.2496 | No | ||
| 51 | PACRG | 13009 | -0.507 | -0.2667 | No | ||
| 52 | PRKCSH | 13182 | -0.555 | -0.2720 | No | ||
| 53 | SPA17 | 13332 | -0.598 | -0.2761 | No | ||
| 54 | TMEM24 | 13590 | -0.672 | -0.2848 | No | ||
| 55 | TUSC3 | 13679 | -0.693 | -0.2857 | No | ||
| 56 | ARID1A | 14066 | -0.796 | -0.2997 | No | ||
| 57 | EMX2 | 14090 | -0.801 | -0.2971 | No | ||
| 58 | NEK2 | 14496 | -0.906 | -0.3115 | No | ||
| 59 | NEDD9 | 14755 | -0.973 | -0.3189 | No | ||
| 60 | ADCK4 | 14842 | -0.996 | -0.3183 | No | ||
| 61 | TRIM39 | 14918 | -1.026 | -0.3171 | No | ||
| 62 | EFNB3 | 14973 | -1.040 | -0.3148 | No | ||
| 63 | LRP2 | 15013 | -1.049 | -0.3118 | No | ||
| 64 | GALNT14 | 15538 | -1.191 | -0.3304 | No | ||
| 65 | STK36 | 15681 | -1.229 | -0.3313 | No | ||
| 66 | CPEB2 | 15832 | -1.269 | -0.3323 | No | ||
| 67 | CARS | 15904 | -1.285 | -0.3297 | No | ||
| 68 | PSCD2 | 15917 | -1.288 | -0.3244 | No | ||
| 69 | NCDN | 16277 | -1.383 | -0.3345 | Yes | ||
| 70 | FGF13 | 16416 | -1.420 | -0.3344 | Yes | ||
| 71 | SNF1LK | 16473 | -1.434 | -0.3304 | Yes | ||
| 72 | TGFB3 | 16663 | -1.485 | -0.3323 | Yes | ||
| 73 | GRM3 | 16820 | -1.532 | -0.3324 | Yes | ||
| 74 | CD44 | 16822 | -1.532 | -0.3255 | Yes | ||
| 75 | FGR | 17077 | -1.610 | -0.3298 | Yes | ||
| 76 | DPAGT1 | 17085 | -1.612 | -0.3228 | Yes | ||
| 77 | CSMD3 | 17238 | -1.661 | -0.3221 | Yes | ||
| 78 | GRID2 | 17434 | -1.729 | -0.3232 | Yes | ||
| 79 | MDH1 | 17511 | -1.754 | -0.3187 | Yes | ||
| 80 | NEDD8 | 17878 | -1.881 | -0.3269 | Yes | ||
| 81 | ITGB8 | 18020 | -1.926 | -0.3245 | Yes | ||
| 82 | MRPL10 | 18031 | -1.930 | -0.3162 | Yes | ||
| 83 | GTF3C4 | 18086 | -1.955 | -0.3097 | Yes | ||
| 84 | ADK | 18271 | -2.024 | -0.3089 | Yes | ||
| 85 | BCL2L1 | 18302 | -2.040 | -0.3010 | Yes | ||
| 86 | VEGF | 18410 | -2.082 | -0.2964 | Yes | ||
| 87 | PTDSS2 | 18974 | -2.325 | -0.3116 | Yes | ||
| 88 | CCDC6 | 19113 | -2.398 | -0.3070 | Yes | ||
| 89 | SREBF1 | 19131 | -2.408 | -0.2968 | Yes | ||
| 90 | CBX6 | 19150 | -2.417 | -0.2866 | Yes | ||
| 91 | MYO1C | 19341 | -2.516 | -0.2838 | Yes | ||
| 92 | AP1M1 | 19404 | -2.554 | -0.2750 | Yes | ||
| 93 | NCOA5 | 19473 | -2.592 | -0.2663 | Yes | ||
| 94 | RNF25 | 19512 | -2.615 | -0.2561 | Yes | ||
| 95 | LSM7 | 19587 | -2.652 | -0.2474 | Yes | ||
| 96 | RFX2 | 19634 | -2.672 | -0.2373 | Yes | ||
| 97 | PLCB3 | 19683 | -2.699 | -0.2272 | Yes | ||
| 98 | ARL4A | 19782 | -2.756 | -0.2191 | Yes | ||
| 99 | MADD | 20356 | -3.143 | -0.2310 | Yes | ||
| 100 | SOCS1 | 20394 | -3.165 | -0.2183 | Yes | ||
| 101 | E2F5 | 20496 | -3.272 | -0.2080 | Yes | ||
| 102 | RNF41 | 20614 | -3.366 | -0.1980 | Yes | ||
| 103 | CBLB | 20738 | -3.491 | -0.1877 | Yes | ||
| 104 | DDX31 | 21246 | -4.252 | -0.1915 | Yes | ||
| 105 | DCTN1 | 21319 | -4.388 | -0.1748 | Yes | ||
| 106 | GMPR2 | 21378 | -4.554 | -0.1567 | Yes | ||
| 107 | DHX30 | 21418 | -4.645 | -0.1373 | Yes | ||
| 108 | YWHAE | 21593 | -5.209 | -0.1215 | Yes | ||
| 109 | CCRK | 21774 | -6.287 | -0.1011 | Yes | ||
| 110 | DNAJC1 | 21786 | -6.436 | -0.0722 | Yes | ||
| 111 | CBX4 | 21833 | -7.190 | -0.0415 | Yes | ||
| 112 | WARS2 | 21907 | -10.243 | 0.0018 | Yes |