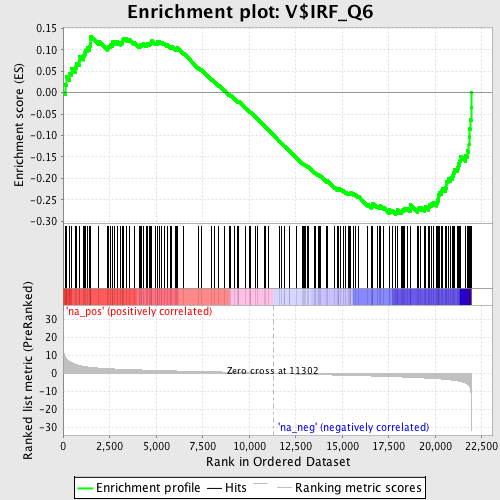
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set01_ATM_minus_versus_SCID |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_neg |
| GeneSet | V$IRF_Q6 |
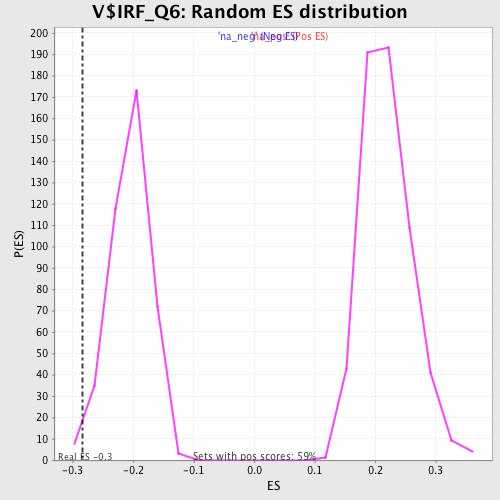
| Enrichment Score (ES) | -0.2841725 |
| Normalized Enrichment Score (NES) | -1.3857746 |
| Nominal p-value | 0.017114915 |
| FDR q-value | 0.37331596 |
| FWER p-Value | 0.997 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | NEDD4 | 109 | 9.109 | 0.0188 | No | ||
| 2 | FCGR2B | 169 | 8.256 | 0.0376 | No | ||
| 3 | EIF4A2 | 356 | 6.429 | 0.0459 | No | ||
| 4 | ETV5 | 461 | 5.827 | 0.0563 | No | ||
| 5 | SYNE2 | 658 | 4.931 | 0.0602 | No | ||
| 6 | B2M | 733 | 4.710 | 0.0691 | No | ||
| 7 | TLK2 | 861 | 4.415 | 0.0748 | No | ||
| 8 | FMR1 | 883 | 4.357 | 0.0852 | No | ||
| 9 | TFDP2 | 1076 | 3.929 | 0.0866 | No | ||
| 10 | CYBB | 1128 | 3.842 | 0.0943 | No | ||
| 11 | CDK6 | 1221 | 3.690 | 0.0997 | No | ||
| 12 | RRBP1 | 1299 | 3.560 | 0.1055 | No | ||
| 13 | MYB | 1437 | 3.386 | 0.1080 | No | ||
| 14 | STAG2 | 1457 | 3.358 | 0.1159 | No | ||
| 15 | PTMA | 1471 | 3.346 | 0.1240 | No | ||
| 16 | TOP1 | 1486 | 3.326 | 0.1321 | No | ||
| 17 | NCF1 | 1919 | 2.923 | 0.1199 | No | ||
| 18 | IFIT2 | 2363 | 2.607 | 0.1064 | No | ||
| 19 | FBXL11 | 2459 | 2.558 | 0.1087 | No | ||
| 20 | MAPK10 | 2538 | 2.517 | 0.1117 | No | ||
| 21 | SEMA6D | 2637 | 2.461 | 0.1136 | No | ||
| 22 | ARHGAP5 | 2666 | 2.449 | 0.1187 | No | ||
| 23 | CNTFR | 2779 | 2.397 | 0.1198 | No | ||
| 24 | BCL11A | 2920 | 2.333 | 0.1195 | No | ||
| 25 | PSMA3 | 3100 | 2.259 | 0.1171 | No | ||
| 26 | ZF | 3177 | 2.229 | 0.1195 | No | ||
| 27 | DUSP10 | 3215 | 2.215 | 0.1236 | No | ||
| 28 | TNFSF13B | 3253 | 2.198 | 0.1276 | No | ||
| 29 | CASK | 3409 | 2.138 | 0.1261 | No | ||
| 30 | PTPRO | 3573 | 2.071 | 0.1240 | No | ||
| 31 | MLLT3 | 3812 | 1.987 | 0.1182 | No | ||
| 32 | HTR2C | 4102 | 1.894 | 0.1099 | No | ||
| 33 | USF1 | 4175 | 1.871 | 0.1115 | No | ||
| 34 | DIO2 | 4237 | 1.852 | 0.1135 | No | ||
| 35 | CCNT2 | 4314 | 1.825 | 0.1148 | No | ||
| 36 | DNASE1L3 | 4469 | 1.777 | 0.1124 | No | ||
| 37 | ZFHX1B | 4532 | 1.759 | 0.1141 | No | ||
| 38 | USP44 | 4634 | 1.720 | 0.1140 | No | ||
| 39 | ADAM15 | 4685 | 1.706 | 0.1161 | No | ||
| 40 | IL18BP | 4724 | 1.690 | 0.1188 | No | ||
| 41 | KLHL13 | 4773 | 1.676 | 0.1210 | No | ||
| 42 | LGP2 | 4978 | 1.612 | 0.1158 | No | ||
| 43 | PDGFRB | 5066 | 1.584 | 0.1160 | No | ||
| 44 | MSX1 | 5068 | 1.584 | 0.1200 | No | ||
| 45 | PRDM16 | 5169 | 1.552 | 0.1195 | No | ||
| 46 | SLC15A3 | 5297 | 1.518 | 0.1176 | No | ||
| 47 | SEMA3A | 5424 | 1.484 | 0.1157 | No | ||
| 48 | LGALS8 | 5597 | 1.435 | 0.1116 | No | ||
| 49 | DLL1 | 5775 | 1.388 | 0.1071 | No | ||
| 50 | GATA6 | 5819 | 1.378 | 0.1087 | No | ||
| 51 | TAPBP | 6035 | 1.327 | 0.1023 | No | ||
| 52 | PLP1 | 6098 | 1.313 | 0.1029 | No | ||
| 53 | TAPBPL | 6124 | 1.304 | 0.1051 | No | ||
| 54 | ISL1 | 6476 | 1.223 | 0.0922 | No | ||
| 55 | PITX2 | 7285 | 1.032 | 0.0578 | No | ||
| 56 | PSMB10 | 7426 | 0.999 | 0.0540 | No | ||
| 57 | LY86 | 7990 | 0.873 | 0.0304 | No | ||
| 58 | CHST1 | 8152 | 0.833 | 0.0252 | No | ||
| 59 | GSDMDC1 | 8371 | 0.789 | 0.0172 | No | ||
| 60 | BBX | 8691 | 0.715 | 0.0045 | No | ||
| 61 | CXCL10 | 8931 | 0.654 | -0.0048 | No | ||
| 62 | EOMES | 9008 | 0.637 | -0.0066 | No | ||
| 63 | ZNF385 | 9184 | 0.597 | -0.0131 | No | ||
| 64 | KYNU | 9385 | 0.551 | -0.0209 | No | ||
| 65 | BTAF1 | 9410 | 0.545 | -0.0205 | No | ||
| 66 | EHD1 | 9426 | 0.542 | -0.0198 | No | ||
| 67 | TCIRG1 | 9796 | 0.438 | -0.0356 | No | ||
| 68 | UBE2L6 | 10012 | 0.379 | -0.0445 | No | ||
| 69 | SLC2A5 | 10066 | 0.365 | -0.0460 | No | ||
| 70 | CDH3 | 10079 | 0.359 | -0.0456 | No | ||
| 71 | SIX1 | 10334 | 0.291 | -0.0565 | No | ||
| 72 | DGKA | 10432 | 0.260 | -0.0603 | No | ||
| 73 | MIA2 | 10819 | 0.146 | -0.0776 | No | ||
| 74 | IFNB1 | 10886 | 0.122 | -0.0803 | No | ||
| 75 | CXCR4 | 11032 | 0.079 | -0.0868 | No | ||
| 76 | TWIST1 | 11637 | -0.106 | -0.1142 | No | ||
| 77 | SLC39A7 | 11743 | -0.141 | -0.1187 | No | ||
| 78 | ZFPM1 | 11885 | -0.181 | -0.1247 | No | ||
| 79 | LGALS9 | 11918 | -0.189 | -0.1256 | No | ||
| 80 | ZADH2 | 12155 | -0.261 | -0.1358 | No | ||
| 81 | RARG | 12185 | -0.271 | -0.1364 | No | ||
| 82 | MYOG | 12556 | -0.381 | -0.1524 | No | ||
| 83 | TCF15 | 12861 | -0.465 | -0.1652 | No | ||
| 84 | ITGB7 | 12915 | -0.481 | -0.1663 | No | ||
| 85 | LZTS2 | 12970 | -0.495 | -0.1675 | No | ||
| 86 | NT5C3 | 13048 | -0.518 | -0.1697 | No | ||
| 87 | F3 | 13148 | -0.546 | -0.1728 | No | ||
| 88 | EGFL6 | 13173 | -0.553 | -0.1725 | No | ||
| 89 | PSMA5 | 13516 | -0.653 | -0.1865 | No | ||
| 90 | CCND1 | 13583 | -0.671 | -0.1878 | No | ||
| 91 | LMO2 | 13701 | -0.700 | -0.1913 | No | ||
| 92 | SNX22 | 13793 | -0.721 | -0.1936 | No | ||
| 93 | BHLHB5 | 13820 | -0.729 | -0.1929 | No | ||
| 94 | ADAM12 | 14172 | -0.823 | -0.2069 | No | ||
| 95 | OSM | 14179 | -0.824 | -0.2050 | No | ||
| 96 | SELL | 14591 | -0.928 | -0.2214 | No | ||
| 97 | IL1RAPL1 | 14730 | -0.965 | -0.2252 | No | ||
| 98 | RORB | 14791 | -0.984 | -0.2254 | No | ||
| 99 | KLF14 | 14806 | -0.988 | -0.2235 | No | ||
| 100 | MAML2 | 14894 | -1.013 | -0.2248 | No | ||
| 101 | GRIA3 | 15046 | -1.059 | -0.2290 | No | ||
| 102 | IFI35 | 15194 | -1.099 | -0.2329 | No | ||
| 103 | BST2 | 15313 | -1.131 | -0.2354 | No | ||
| 104 | TBX1 | 15374 | -1.146 | -0.2351 | No | ||
| 105 | MUSK | 15439 | -1.165 | -0.2350 | No | ||
| 106 | CPEB4 | 15466 | -1.173 | -0.2332 | No | ||
| 107 | ACSL5 | 15593 | -1.206 | -0.2358 | No | ||
| 108 | KCNQ4 | 15710 | -1.234 | -0.2379 | No | ||
| 109 | LSP1 | 15878 | -1.279 | -0.2422 | No | ||
| 110 | PCDH7 | 16364 | -1.406 | -0.2608 | No | ||
| 111 | FGF9 | 16548 | -1.454 | -0.2654 | No | ||
| 112 | HAPLN1 | 16601 | -1.469 | -0.2640 | No | ||
| 113 | IFI44 | 16611 | -1.471 | -0.2606 | No | ||
| 114 | BCOR | 16641 | -1.479 | -0.2580 | No | ||
| 115 | MS4A1 | 16879 | -1.547 | -0.2649 | No | ||
| 116 | PCDHGC3 | 16982 | -1.579 | -0.2654 | No | ||
| 117 | EREG | 17031 | -1.594 | -0.2635 | No | ||
| 118 | PURA | 17225 | -1.659 | -0.2680 | No | ||
| 119 | STAT6 | 17516 | -1.755 | -0.2767 | No | ||
| 120 | UPF2 | 17531 | -1.761 | -0.2728 | No | ||
| 121 | DCAMKL1 | 17672 | -1.809 | -0.2745 | No | ||
| 122 | SLC40A1 | 17884 | -1.883 | -0.2793 | Yes | ||
| 123 | MOV10 | 17967 | -1.908 | -0.2780 | Yes | ||
| 124 | ECM2 | 17973 | -1.911 | -0.2733 | Yes | ||
| 125 | NRP1 | 18183 | -1.990 | -0.2777 | Yes | ||
| 126 | PLXNC1 | 18210 | -2.000 | -0.2737 | Yes | ||
| 127 | HOXB3 | 18298 | -2.036 | -0.2723 | Yes | ||
| 128 | TCF7L2 | 18354 | -2.060 | -0.2695 | Yes | ||
| 129 | CD80 | 18489 | -2.115 | -0.2701 | Yes | ||
| 130 | HOXB4 | 18643 | -2.180 | -0.2714 | Yes | ||
| 131 | RAPGEF6 | 18652 | -2.185 | -0.2661 | Yes | ||
| 132 | TLR7 | 18655 | -2.186 | -0.2605 | Yes | ||
| 133 | ESR1 | 19068 | -2.374 | -0.2732 | Yes | ||
| 134 | CCDC6 | 19113 | -2.398 | -0.2690 | Yes | ||
| 135 | PRKACA | 19225 | -2.460 | -0.2677 | Yes | ||
| 136 | PSMB8 | 19413 | -2.558 | -0.2696 | Yes | ||
| 137 | NPEPPS | 19451 | -2.574 | -0.2645 | Yes | ||
| 138 | ARHGEF6 | 19656 | -2.685 | -0.2669 | Yes | ||
| 139 | NLGN2 | 19703 | -2.712 | -0.2619 | Yes | ||
| 140 | SLC11A2 | 19807 | -2.767 | -0.2594 | Yes | ||
| 141 | PRKAG1 | 19884 | -2.818 | -0.2556 | Yes | ||
| 142 | HPCAL1 | 20078 | -2.948 | -0.2567 | Yes | ||
| 143 | ARF3 | 20104 | -2.968 | -0.2501 | Yes | ||
| 144 | RBMS2 | 20172 | -3.013 | -0.2453 | Yes | ||
| 145 | SFRS12 | 20190 | -3.022 | -0.2382 | Yes | ||
| 146 | ADAR | 20231 | -3.048 | -0.2321 | Yes | ||
| 147 | PTK2 | 20343 | -3.132 | -0.2290 | Yes | ||
| 148 | SOCS1 | 20394 | -3.165 | -0.2231 | Yes | ||
| 149 | LGI1 | 20542 | -3.300 | -0.2212 | Yes | ||
| 150 | ISGF3G | 20598 | -3.358 | -0.2150 | Yes | ||
| 151 | ETV6 | 20603 | -3.360 | -0.2064 | Yes | ||
| 152 | PIP5K2B | 20683 | -3.430 | -0.2010 | Yes | ||
| 153 | DAPP1 | 20841 | -3.613 | -0.1988 | Yes | ||
| 154 | ZBP1 | 20906 | -3.691 | -0.1921 | Yes | ||
| 155 | RTN4 | 20996 | -3.813 | -0.1863 | Yes | ||
| 156 | MAP3K11 | 21046 | -3.888 | -0.1784 | Yes | ||
| 157 | GABARAP | 21171 | -4.086 | -0.1734 | Yes | ||
| 158 | GRIPAP1 | 21231 | -4.207 | -0.1651 | Yes | ||
| 159 | FUT8 | 21323 | -4.397 | -0.1578 | Yes | ||
| 160 | PRKD2 | 21372 | -4.534 | -0.1482 | Yes | ||
| 161 | FLI1 | 21627 | -5.352 | -0.1459 | Yes | ||
| 162 | HIPK1 | 21734 | -5.953 | -0.1352 | Yes | ||
| 163 | NFATC1 | 21808 | -6.771 | -0.1209 | Yes | ||
| 164 | CBX4 | 21833 | -7.190 | -0.1032 | Yes | ||
| 165 | IRF2 | 21846 | -7.452 | -0.0843 | Yes | ||
| 166 | EXT1 | 21887 | -8.877 | -0.0630 | Yes | ||
| 167 | CASP7 | 21924 | -11.616 | -0.0343 | Yes | ||
| 168 | SERHL | 21937 | -13.521 | 0.0005 | Yes |