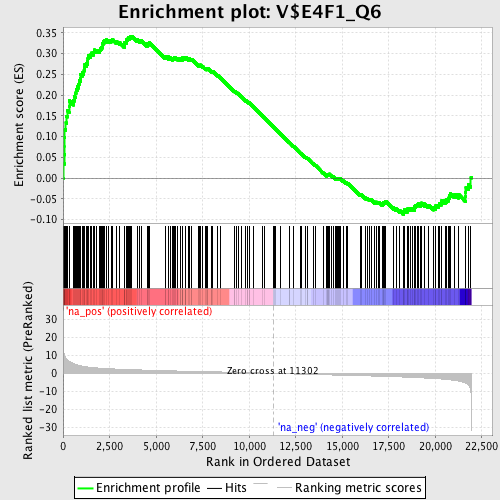
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set01_ATM_minus_versus_SCID |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_pos |
| GeneSet | V$E4F1_Q6 |
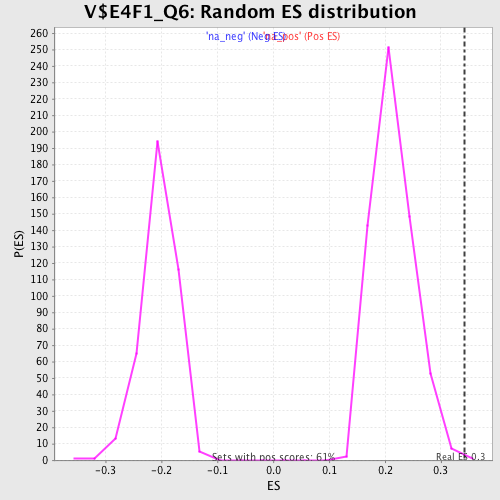
| Enrichment Score (ES) | 0.34209976 |
| Normalized Enrichment Score (NES) | 1.595875 |
| Nominal p-value | 0.0016528926 |
| FDR q-value | 0.092808865 |
| FWER p-Value | 0.801 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | SMARCE1 | 7 | 16.881 | 0.0360 | Yes | ||
| 2 | HNRPA2B1 | 59 | 10.466 | 0.0562 | Yes | ||
| 3 | DHX36 | 74 | 10.011 | 0.0771 | Yes | ||
| 4 | ETF1 | 77 | 9.911 | 0.0984 | Yes | ||
| 5 | DDX3X | 100 | 9.225 | 0.1172 | Yes | ||
| 6 | PRG1 | 154 | 8.424 | 0.1329 | Yes | ||
| 7 | PSMD12 | 183 | 8.008 | 0.1489 | Yes | ||
| 8 | ZNF24 | 222 | 7.510 | 0.1633 | Yes | ||
| 9 | SEC63 | 323 | 6.661 | 0.1731 | Yes | ||
| 10 | NINJ1 | 333 | 6.588 | 0.1868 | Yes | ||
| 11 | RPS6KA3 | 571 | 5.239 | 0.1872 | Yes | ||
| 12 | YWHAG | 608 | 5.096 | 0.1965 | Yes | ||
| 13 | TNFAIP8 | 654 | 4.932 | 0.2051 | Yes | ||
| 14 | SLC31A1 | 695 | 4.828 | 0.2136 | Yes | ||
| 15 | STAG1 | 770 | 4.621 | 0.2202 | Yes | ||
| 16 | SFRS1 | 843 | 4.448 | 0.2264 | Yes | ||
| 17 | RAB6A | 862 | 4.413 | 0.2351 | Yes | ||
| 18 | ELOVL5 | 932 | 4.218 | 0.2410 | Yes | ||
| 19 | ATP2A2 | 947 | 4.197 | 0.2494 | Yes | ||
| 20 | PTP4A1 | 1028 | 4.027 | 0.2544 | Yes | ||
| 21 | ATP6V0C | 1121 | 3.858 | 0.2585 | Yes | ||
| 22 | UBQLN2 | 1133 | 3.831 | 0.2662 | Yes | ||
| 23 | GNAS | 1168 | 3.771 | 0.2728 | Yes | ||
| 24 | PPP2R2A | 1272 | 3.605 | 0.2758 | Yes | ||
| 25 | ARF4 | 1319 | 3.529 | 0.2813 | Yes | ||
| 26 | KDELR3 | 1333 | 3.514 | 0.2883 | Yes | ||
| 27 | MORF4L2 | 1347 | 3.499 | 0.2952 | Yes | ||
| 28 | PTMA | 1471 | 3.346 | 0.2968 | Yes | ||
| 29 | HMGB1 | 1523 | 3.287 | 0.3015 | Yes | ||
| 30 | MASTL | 1636 | 3.169 | 0.3032 | Yes | ||
| 31 | PLK4 | 1661 | 3.143 | 0.3088 | Yes | ||
| 32 | IKBKB | 1816 | 3.007 | 0.3082 | Yes | ||
| 33 | CCNI | 1957 | 2.878 | 0.3080 | Yes | ||
| 34 | WBP5 | 1992 | 2.855 | 0.3126 | Yes | ||
| 35 | OSR1 | 2071 | 2.789 | 0.3150 | Yes | ||
| 36 | GOLGA5 | 2108 | 2.763 | 0.3193 | Yes | ||
| 37 | EGR2 | 2110 | 2.763 | 0.3252 | Yes | ||
| 38 | PKP4 | 2161 | 2.728 | 0.3288 | Yes | ||
| 39 | ALS2CR13 | 2234 | 2.681 | 0.3312 | Yes | ||
| 40 | ATF1 | 2314 | 2.637 | 0.3333 | Yes | ||
| 41 | FBXL11 | 2459 | 2.558 | 0.3322 | Yes | ||
| 42 | EGR4 | 2573 | 2.496 | 0.3323 | Yes | ||
| 43 | FKBP7 | 2640 | 2.459 | 0.3346 | Yes | ||
| 44 | HIST1H4H | 2861 | 2.362 | 0.3296 | Yes | ||
| 45 | GTF2A1 | 3046 | 2.279 | 0.3260 | Yes | ||
| 46 | SLC18A2 | 3296 | 2.183 | 0.3193 | Yes | ||
| 47 | PSME3 | 3312 | 2.176 | 0.3233 | Yes | ||
| 48 | NR4A1 | 3314 | 2.175 | 0.3279 | Yes | ||
| 49 | EHD4 | 3390 | 2.144 | 0.3291 | Yes | ||
| 50 | UBAP1 | 3397 | 2.142 | 0.3334 | Yes | ||
| 51 | CCBL1 | 3458 | 2.123 | 0.3352 | Yes | ||
| 52 | MRC2 | 3486 | 2.108 | 0.3385 | Yes | ||
| 53 | CREB1 | 3579 | 2.070 | 0.3387 | Yes | ||
| 54 | NR3C1 | 3635 | 2.049 | 0.3406 | Yes | ||
| 55 | ADNP | 3699 | 2.027 | 0.3421 | Yes | ||
| 56 | PRR3 | 3975 | 1.937 | 0.3336 | No | ||
| 57 | EDG5 | 4111 | 1.891 | 0.3315 | No | ||
| 58 | CLDN3 | 4193 | 1.865 | 0.3318 | No | ||
| 59 | ALDOC | 4516 | 1.764 | 0.3208 | No | ||
| 60 | ZFHX1B | 4532 | 1.759 | 0.3239 | No | ||
| 61 | FOSB | 4608 | 1.729 | 0.3242 | No | ||
| 62 | HS6ST2 | 4643 | 1.715 | 0.3263 | No | ||
| 63 | LENG9 | 5480 | 1.465 | 0.2910 | No | ||
| 64 | RHOBTB1 | 5509 | 1.457 | 0.2929 | No | ||
| 65 | TRIB1 | 5655 | 1.422 | 0.2892 | No | ||
| 66 | RAB2 | 5659 | 1.420 | 0.2922 | No | ||
| 67 | RUSC1 | 5778 | 1.387 | 0.2897 | No | ||
| 68 | CD160 | 5901 | 1.356 | 0.2870 | No | ||
| 69 | CD2AP | 5915 | 1.352 | 0.2894 | No | ||
| 70 | TBRG4 | 5962 | 1.343 | 0.2901 | No | ||
| 71 | AGPAT1 | 6032 | 1.327 | 0.2898 | No | ||
| 72 | SLC36A3 | 6150 | 1.297 | 0.2872 | No | ||
| 73 | CXCL16 | 6167 | 1.291 | 0.2893 | No | ||
| 74 | NUMBL | 6283 | 1.265 | 0.2867 | No | ||
| 75 | GRIN2B | 6287 | 1.264 | 0.2893 | No | ||
| 76 | FLRT3 | 6402 | 1.239 | 0.2867 | No | ||
| 77 | GEM | 6426 | 1.234 | 0.2883 | No | ||
| 78 | SLC20A2 | 6432 | 1.233 | 0.2907 | No | ||
| 79 | RBBP8 | 6555 | 1.206 | 0.2877 | No | ||
| 80 | SATB2 | 6562 | 1.205 | 0.2900 | No | ||
| 81 | TAF10 | 6585 | 1.199 | 0.2916 | No | ||
| 82 | SLC25A25 | 6758 | 1.156 | 0.2862 | No | ||
| 83 | ZMYND15 | 6770 | 1.155 | 0.2882 | No | ||
| 84 | FOXA1 | 6873 | 1.128 | 0.2859 | No | ||
| 85 | PDE7A | 6905 | 1.121 | 0.2869 | No | ||
| 86 | PITX2 | 7285 | 1.032 | 0.2717 | No | ||
| 87 | PPM2C | 7340 | 1.019 | 0.2714 | No | ||
| 88 | EIF5A | 7362 | 1.015 | 0.2726 | No | ||
| 89 | CLDN7 | 7497 | 0.987 | 0.2686 | No | ||
| 90 | SCG2 | 7654 | 0.948 | 0.2635 | No | ||
| 91 | SH2D2A | 7721 | 0.936 | 0.2625 | No | ||
| 92 | UFC1 | 7734 | 0.933 | 0.2639 | No | ||
| 93 | CX36 | 7750 | 0.931 | 0.2652 | No | ||
| 94 | SHC3 | 7979 | 0.876 | 0.2566 | No | ||
| 95 | SCAMP5 | 7996 | 0.872 | 0.2578 | No | ||
| 96 | KIRREL3 | 8051 | 0.858 | 0.2571 | No | ||
| 97 | FZD7 | 8301 | 0.804 | 0.2474 | No | ||
| 98 | SYNGR3 | 8469 | 0.767 | 0.2414 | No | ||
| 99 | TRPC1 | 9209 | 0.590 | 0.2087 | No | ||
| 100 | CSTF3 | 9224 | 0.588 | 0.2093 | No | ||
| 101 | PPP2R2B | 9329 | 0.564 | 0.2057 | No | ||
| 102 | HOXC10 | 9411 | 0.544 | 0.2032 | No | ||
| 103 | FSTL5 | 9582 | 0.499 | 0.1965 | No | ||
| 104 | DDX51 | 9824 | 0.430 | 0.1863 | No | ||
| 105 | FOXD3 | 9909 | 0.406 | 0.1833 | No | ||
| 106 | MAP1LC3A | 9925 | 0.403 | 0.1835 | No | ||
| 107 | IRX6 | 9998 | 0.382 | 0.1810 | No | ||
| 108 | CHGB | 10001 | 0.381 | 0.1817 | No | ||
| 109 | GPR3 | 10203 | 0.326 | 0.1732 | No | ||
| 110 | SLC30A5 | 10688 | 0.189 | 0.1513 | No | ||
| 111 | NKX2-2 | 10827 | 0.145 | 0.1453 | No | ||
| 112 | NEUROD6 | 11288 | 0.003 | 0.1242 | No | ||
| 113 | GOLGA2 | 11362 | -0.021 | 0.1209 | No | ||
| 114 | ID1 | 11419 | -0.040 | 0.1184 | No | ||
| 115 | PNMA3 | 11694 | -0.125 | 0.1060 | No | ||
| 116 | ZADH2 | 12155 | -0.261 | 0.0855 | No | ||
| 117 | CALM2 | 12170 | -0.266 | 0.0854 | No | ||
| 118 | NAP1L2 | 12379 | -0.325 | 0.0765 | No | ||
| 119 | ZNF462 | 12754 | -0.438 | 0.0603 | No | ||
| 120 | FAU | 12788 | -0.448 | 0.0597 | No | ||
| 121 | DUSP1 | 13018 | -0.508 | 0.0503 | No | ||
| 122 | KCNA2 | 13111 | -0.535 | 0.0472 | No | ||
| 123 | SMARCA1 | 13129 | -0.539 | 0.0476 | No | ||
| 124 | RFX5 | 13427 | -0.629 | 0.0353 | No | ||
| 125 | DACT1 | 13548 | -0.662 | 0.0312 | No | ||
| 126 | MCAM | 13993 | -0.776 | 0.0124 | No | ||
| 127 | WNT10A | 14165 | -0.821 | 0.0064 | No | ||
| 128 | RPL41 | 14180 | -0.824 | 0.0075 | No | ||
| 129 | MAFF | 14202 | -0.827 | 0.0083 | No | ||
| 130 | CKB | 14256 | -0.840 | 0.0077 | No | ||
| 131 | ZBTB4 | 14269 | -0.843 | 0.0089 | No | ||
| 132 | EGR3 | 14289 | -0.850 | 0.0099 | No | ||
| 133 | PDLIM3 | 14432 | -0.886 | 0.0053 | No | ||
| 134 | SSTR2 | 14522 | -0.914 | 0.0031 | No | ||
| 135 | MAP1A | 14655 | -0.945 | -0.0009 | No | ||
| 136 | PPP6C | 14680 | -0.954 | 0.0001 | No | ||
| 137 | NEDD9 | 14755 | -0.973 | -0.0012 | No | ||
| 138 | TBX20 | 14780 | -0.980 | -0.0002 | No | ||
| 139 | RNF5 | 14832 | -0.994 | -0.0004 | No | ||
| 140 | EIF4A1 | 14880 | -1.009 | -0.0004 | No | ||
| 141 | ERF | 15068 | -1.064 | -0.0067 | No | ||
| 142 | CBX3 | 15206 | -1.103 | -0.0107 | No | ||
| 143 | SOX9 | 15290 | -1.126 | -0.0121 | No | ||
| 144 | TSPYL5 | 15964 | -1.301 | -0.0402 | No | ||
| 145 | MAF | 16023 | -1.316 | -0.0400 | No | ||
| 146 | SLC39A13 | 16254 | -1.376 | -0.0476 | No | ||
| 147 | GNG4 | 16342 | -1.401 | -0.0486 | No | ||
| 148 | SNF1LK | 16473 | -1.434 | -0.0515 | No | ||
| 149 | FGF9 | 16548 | -1.454 | -0.0518 | No | ||
| 150 | NOL4 | 16745 | -1.511 | -0.0575 | No | ||
| 151 | GRM3 | 16820 | -1.532 | -0.0576 | No | ||
| 152 | LTBP1 | 16926 | -1.562 | -0.0591 | No | ||
| 153 | NR6A1 | 16980 | -1.579 | -0.0581 | No | ||
| 154 | USP48 | 17178 | -1.641 | -0.0637 | No | ||
| 155 | SRPR | 17204 | -1.650 | -0.0613 | No | ||
| 156 | ADAMTS3 | 17213 | -1.654 | -0.0581 | No | ||
| 157 | C1QTNF7 | 17284 | -1.677 | -0.0577 | No | ||
| 158 | MYST1 | 17322 | -1.690 | -0.0557 | No | ||
| 159 | ANK2 | 17750 | -1.836 | -0.0714 | No | ||
| 160 | CRY2 | 17901 | -1.890 | -0.0742 | No | ||
| 161 | PPARGC1A | 18084 | -1.952 | -0.0784 | No | ||
| 162 | NEK1 | 18288 | -2.029 | -0.0834 | No | ||
| 163 | HOXB3 | 18298 | -2.036 | -0.0794 | No | ||
| 164 | GRK6 | 18317 | -2.045 | -0.0758 | No | ||
| 165 | ARG2 | 18482 | -2.112 | -0.0788 | No | ||
| 166 | ATP10D | 18485 | -2.113 | -0.0744 | No | ||
| 167 | KCNA6 | 18564 | -2.143 | -0.0733 | No | ||
| 168 | OSBP | 18675 | -2.191 | -0.0737 | No | ||
| 169 | FGF14 | 18792 | -2.246 | -0.0742 | No | ||
| 170 | NR4A2 | 18880 | -2.284 | -0.0733 | No | ||
| 171 | EPC1 | 18893 | -2.291 | -0.0689 | No | ||
| 172 | PTOV1 | 18927 | -2.304 | -0.0654 | No | ||
| 173 | HIC2 | 19020 | -2.347 | -0.0646 | No | ||
| 174 | BRAF | 19077 | -2.378 | -0.0621 | No | ||
| 175 | RFX1 | 19204 | -2.448 | -0.0626 | No | ||
| 176 | CALCRL | 19265 | -2.480 | -0.0600 | No | ||
| 177 | PURB | 19434 | -2.565 | -0.0622 | No | ||
| 178 | SACM1L | 19647 | -2.681 | -0.0662 | No | ||
| 179 | PRKAG1 | 19884 | -2.818 | -0.0710 | No | ||
| 180 | NDUFB2 | 20003 | -2.899 | -0.0701 | No | ||
| 181 | CALD1 | 20032 | -2.916 | -0.0651 | No | ||
| 182 | RBMS2 | 20172 | -3.013 | -0.0651 | No | ||
| 183 | CBX8 | 20216 | -3.039 | -0.0605 | No | ||
| 184 | RANBP2 | 20315 | -3.112 | -0.0583 | No | ||
| 185 | CYR61 | 20353 | -3.140 | -0.0532 | No | ||
| 186 | POLR2A | 20527 | -3.291 | -0.0541 | No | ||
| 187 | BCL2L13 | 20616 | -3.369 | -0.0509 | No | ||
| 188 | GAK | 20716 | -3.463 | -0.0480 | No | ||
| 189 | NR4A3 | 20751 | -3.509 | -0.0420 | No | ||
| 190 | DNM3 | 20826 | -3.594 | -0.0377 | No | ||
| 191 | MAP3K13 | 21048 | -3.890 | -0.0394 | No | ||
| 192 | RAB1A | 21240 | -4.235 | -0.0391 | No | ||
| 193 | MRPL49 | 21618 | -5.304 | -0.0450 | No | ||
| 194 | PAFAH1B1 | 21628 | -5.352 | -0.0339 | No | ||
| 195 | TLE3 | 21646 | -5.426 | -0.0230 | No | ||
| 196 | PCQAP | 21771 | -6.214 | -0.0153 | No | ||
| 197 | ABCF3 | 21915 | -10.854 | 0.0015 | No |