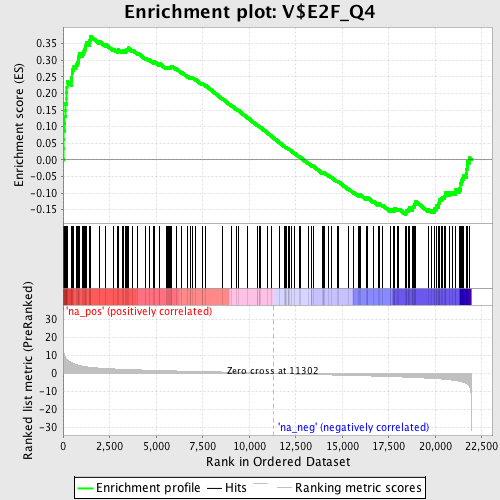
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set01_ATM_minus_versus_SCID |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_pos |
| GeneSet | V$E2F_Q4 |
| Enrichment Score (ES) | 0.37243223 |
| Normalized Enrichment Score (NES) | 1.6939934 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.058353428 |
| FWER p-Value | 0.442 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | SFRS2 | 12 | 15.098 | 0.0330 | Yes | ||
| 2 | SFRS10 | 19 | 13.401 | 0.0625 | Yes | ||
| 3 | NCL | 36 | 11.624 | 0.0876 | Yes | ||
| 4 | EZH2 | 58 | 10.502 | 0.1099 | Yes | ||
| 5 | MCM4 | 79 | 9.758 | 0.1307 | Yes | ||
| 6 | GPRC5B | 115 | 9.024 | 0.1491 | Yes | ||
| 7 | GSPT1 | 122 | 8.818 | 0.1684 | Yes | ||
| 8 | NASP | 168 | 8.269 | 0.1847 | Yes | ||
| 9 | ING3 | 170 | 8.243 | 0.2030 | Yes | ||
| 10 | SFRS7 | 206 | 7.661 | 0.2184 | Yes | ||
| 11 | HNRPA1 | 209 | 7.587 | 0.2351 | Yes | ||
| 12 | PRPS1 | 441 | 5.930 | 0.2377 | Yes | ||
| 13 | KPNB1 | 470 | 5.778 | 0.2493 | Yes | ||
| 14 | WHSC1L1 | 477 | 5.742 | 0.2617 | Yes | ||
| 15 | NUP153 | 524 | 5.472 | 0.2718 | Yes | ||
| 16 | NOLC1 | 581 | 5.207 | 0.2808 | Yes | ||
| 17 | PB1 | 693 | 4.835 | 0.2864 | Yes | ||
| 18 | STAG1 | 770 | 4.621 | 0.2932 | Yes | ||
| 19 | TOPBP1 | 811 | 4.527 | 0.3014 | Yes | ||
| 20 | SFRS1 | 843 | 4.448 | 0.3099 | Yes | ||
| 21 | HNRPR | 858 | 4.419 | 0.3190 | Yes | ||
| 22 | RPS6KA5 | 1025 | 4.034 | 0.3204 | Yes | ||
| 23 | E2F3 | 1108 | 3.880 | 0.3252 | Yes | ||
| 24 | IPO7 | 1127 | 3.845 | 0.3330 | Yes | ||
| 25 | CTNND2 | 1180 | 3.742 | 0.3389 | Yes | ||
| 26 | SLCO3A1 | 1216 | 3.697 | 0.3455 | Yes | ||
| 27 | AP4M1 | 1247 | 3.641 | 0.3522 | Yes | ||
| 28 | CRK | 1421 | 3.408 | 0.3518 | Yes | ||
| 29 | POLD3 | 1430 | 3.399 | 0.3590 | Yes | ||
| 30 | STAG2 | 1457 | 3.358 | 0.3653 | Yes | ||
| 31 | RBL1 | 1464 | 3.353 | 0.3724 | Yes | ||
| 32 | UFD1L | 1946 | 2.891 | 0.3568 | No | ||
| 33 | HTF9C | 2271 | 2.662 | 0.3478 | No | ||
| 34 | PCNA | 2701 | 2.431 | 0.3335 | No | ||
| 35 | IL4I1 | 2918 | 2.334 | 0.3288 | No | ||
| 36 | JPH1 | 2984 | 2.308 | 0.3309 | No | ||
| 37 | INSM1 | 3181 | 2.227 | 0.3269 | No | ||
| 38 | RANBP1 | 3228 | 2.211 | 0.3297 | No | ||
| 39 | ARHGAP11A | 3353 | 2.160 | 0.3288 | No | ||
| 40 | HNRPD | 3388 | 2.146 | 0.3320 | No | ||
| 41 | UGCGL1 | 3475 | 2.113 | 0.3327 | No | ||
| 42 | ABI2 | 3488 | 2.107 | 0.3369 | No | ||
| 43 | AK2 | 3741 | 2.013 | 0.3298 | No | ||
| 44 | ASXL2 | 4020 | 1.918 | 0.3213 | No | ||
| 45 | MSH2 | 4451 | 1.783 | 0.3055 | No | ||
| 46 | ARHGAP6 | 4616 | 1.725 | 0.3018 | No | ||
| 47 | ID3 | 4852 | 1.653 | 0.2947 | No | ||
| 48 | PODN | 4914 | 1.630 | 0.2955 | No | ||
| 49 | FBXO5 | 5171 | 1.551 | 0.2872 | No | ||
| 50 | MCM6 | 5172 | 1.550 | 0.2906 | No | ||
| 51 | PAX6 | 5532 | 1.451 | 0.2774 | No | ||
| 52 | PHF5A | 5598 | 1.435 | 0.2776 | No | ||
| 53 | E2F1 | 5666 | 1.419 | 0.2776 | No | ||
| 54 | ACBD6 | 5734 | 1.399 | 0.2777 | No | ||
| 55 | WEE1 | 5782 | 1.386 | 0.2786 | No | ||
| 56 | PHF15 | 5799 | 1.382 | 0.2809 | No | ||
| 57 | MCM8 | 5839 | 1.373 | 0.2822 | No | ||
| 58 | KBTBD7 | 6080 | 1.316 | 0.2741 | No | ||
| 59 | TMPO | 6375 | 1.245 | 0.2634 | No | ||
| 60 | PPM1D | 6687 | 1.174 | 0.2517 | No | ||
| 61 | PIM1 | 6823 | 1.141 | 0.2480 | No | ||
| 62 | CSPG2 | 6866 | 1.130 | 0.2486 | No | ||
| 63 | DMD | 6960 | 1.109 | 0.2468 | No | ||
| 64 | HCN3 | 7116 | 1.070 | 0.2421 | No | ||
| 65 | KCND2 | 7474 | 0.991 | 0.2279 | No | ||
| 66 | MCM3 | 7488 | 0.988 | 0.2295 | No | ||
| 67 | DDB2 | 7671 | 0.946 | 0.2232 | No | ||
| 68 | MRPL40 | 8576 | 0.740 | 0.1834 | No | ||
| 69 | ATF5 | 9046 | 0.628 | 0.1632 | No | ||
| 70 | RAD51 | 9305 | 0.569 | 0.1526 | No | ||
| 71 | HOXC10 | 9411 | 0.544 | 0.1490 | No | ||
| 72 | UXT | 9922 | 0.404 | 0.1265 | No | ||
| 73 | HMGN2 | 10426 | 0.262 | 0.1040 | No | ||
| 74 | UNG | 10452 | 0.255 | 0.1034 | No | ||
| 75 | RQCD1 | 10542 | 0.231 | 0.0999 | No | ||
| 76 | SNRPD1 | 10547 | 0.229 | 0.1002 | No | ||
| 77 | RAB11B | 10622 | 0.207 | 0.0972 | No | ||
| 78 | TBX6 | 11008 | 0.086 | 0.0798 | No | ||
| 79 | STMN1 | 11211 | 0.027 | 0.0705 | No | ||
| 80 | HS6ST3 | 11600 | -0.096 | 0.0529 | No | ||
| 81 | DNAJC9 | 11901 | -0.186 | 0.0396 | No | ||
| 82 | ALDH6A1 | 11952 | -0.198 | 0.0377 | No | ||
| 83 | PCSK1 | 11955 | -0.199 | 0.0381 | No | ||
| 84 | FMO4 | 12029 | -0.219 | 0.0352 | No | ||
| 85 | PRKDC | 12100 | -0.245 | 0.0325 | No | ||
| 86 | CORT | 12105 | -0.247 | 0.0329 | No | ||
| 87 | MCM7 | 12175 | -0.266 | 0.0303 | No | ||
| 88 | EED | 12272 | -0.293 | 0.0266 | No | ||
| 89 | WNT3 | 12450 | -0.349 | 0.0192 | No | ||
| 90 | PAQR4 | 12691 | -0.419 | 0.0091 | No | ||
| 91 | HNRPUL1 | 12768 | -0.442 | 0.0066 | No | ||
| 92 | RRM2 | 13177 | -0.554 | -0.0109 | No | ||
| 93 | ADAMTS2 | 13355 | -0.606 | -0.0176 | No | ||
| 94 | SLC6A4 | 13358 | -0.607 | -0.0164 | No | ||
| 95 | RHD | 13435 | -0.630 | -0.0185 | No | ||
| 96 | CASP8AP2 | 13952 | -0.766 | -0.0405 | No | ||
| 97 | TREX2 | 13981 | -0.774 | -0.0400 | No | ||
| 98 | EPHB1 | 14001 | -0.779 | -0.0392 | No | ||
| 99 | ZCCHC8 | 14040 | -0.789 | -0.0392 | No | ||
| 100 | ZBTB4 | 14269 | -0.843 | -0.0478 | No | ||
| 101 | CTCF | 14413 | -0.882 | -0.0524 | No | ||
| 102 | RASAL2 | 14720 | -0.962 | -0.0643 | No | ||
| 103 | ILF3 | 14779 | -0.980 | -0.0648 | No | ||
| 104 | ARID4A | 15331 | -1.135 | -0.0875 | No | ||
| 105 | FANCD2 | 15609 | -1.209 | -0.0976 | No | ||
| 106 | PHC1 | 15865 | -1.276 | -0.1064 | No | ||
| 107 | SYNGR4 | 15905 | -1.285 | -0.1054 | No | ||
| 108 | DCK | 15954 | -1.299 | -0.1047 | No | ||
| 109 | CDT1 | 16293 | -1.388 | -0.1171 | No | ||
| 110 | GRIA4 | 16358 | -1.406 | -0.1169 | No | ||
| 111 | GABRB3 | 16371 | -1.408 | -0.1144 | No | ||
| 112 | FHOD1 | 16660 | -1.484 | -0.1243 | No | ||
| 113 | SEMA5A | 16919 | -1.559 | -0.1327 | No | ||
| 114 | SLC9A5 | 16988 | -1.582 | -0.1323 | No | ||
| 115 | SOAT1 | 17153 | -1.633 | -0.1362 | No | ||
| 116 | MCM2 | 17573 | -1.777 | -0.1515 | No | ||
| 117 | NRP2 | 17597 | -1.784 | -0.1486 | No | ||
| 118 | SLC38A1 | 17749 | -1.836 | -0.1514 | No | ||
| 119 | POLE2 | 17811 | -1.853 | -0.1501 | No | ||
| 120 | EFNA5 | 17815 | -1.855 | -0.1461 | No | ||
| 121 | CCNT1 | 17975 | -1.912 | -0.1492 | No | ||
| 122 | NELL2 | 18047 | -1.941 | -0.1481 | No | ||
| 123 | PKMYT1 | 18405 | -2.081 | -0.1599 | No | ||
| 124 | CLSPN | 18429 | -2.090 | -0.1563 | No | ||
| 125 | CDC25A | 18430 | -2.090 | -0.1517 | No | ||
| 126 | BRMS1L | 18546 | -2.138 | -0.1522 | No | ||
| 127 | KCNA6 | 18564 | -2.143 | -0.1482 | No | ||
| 128 | PHF13 | 18585 | -2.149 | -0.1444 | No | ||
| 129 | USP37 | 18762 | -2.228 | -0.1475 | No | ||
| 130 | POLD1 | 18799 | -2.249 | -0.1442 | No | ||
| 131 | DDX17 | 18811 | -2.253 | -0.1397 | No | ||
| 132 | CDC45L | 18866 | -2.277 | -0.1371 | No | ||
| 133 | EPC1 | 18893 | -2.291 | -0.1332 | No | ||
| 134 | DNAJC5G | 18934 | -2.306 | -0.1299 | No | ||
| 135 | MYH10 | 18956 | -2.317 | -0.1257 | No | ||
| 136 | CDC6 | 19624 | -2.669 | -0.1504 | No | ||
| 137 | POLA2 | 19818 | -2.775 | -0.1531 | No | ||
| 138 | NUP62 | 19937 | -2.850 | -0.1522 | No | ||
| 139 | TYRO3 | 19968 | -2.870 | -0.1472 | No | ||
| 140 | ACO2 | 20054 | -2.927 | -0.1446 | No | ||
| 141 | GMNN | 20067 | -2.936 | -0.1386 | No | ||
| 142 | PPP1CC | 20164 | -3.005 | -0.1364 | No | ||
| 143 | SFRS12 | 20190 | -3.022 | -0.1308 | No | ||
| 144 | POLE4 | 20237 | -3.051 | -0.1261 | No | ||
| 145 | CBX5 | 20242 | -3.054 | -0.1195 | No | ||
| 146 | SUMO1 | 20323 | -3.118 | -0.1163 | No | ||
| 147 | ATAD2 | 20395 | -3.167 | -0.1125 | No | ||
| 148 | DNAJC11 | 20482 | -3.255 | -0.1092 | No | ||
| 149 | POLR2A | 20527 | -3.291 | -0.1039 | No | ||
| 150 | STK35 | 20560 | -3.313 | -0.0980 | No | ||
| 151 | PVRL1 | 20744 | -3.503 | -0.0987 | No | ||
| 152 | CHD2 | 20895 | -3.680 | -0.0974 | No | ||
| 153 | HIRA | 21077 | -3.929 | -0.0970 | No | ||
| 154 | H2AFZ | 21106 | -3.980 | -0.0894 | No | ||
| 155 | PPP1R8 | 21275 | -4.306 | -0.0875 | No | ||
| 156 | RAVER1 | 21359 | -4.509 | -0.0813 | No | ||
| 157 | E2F7 | 21360 | -4.512 | -0.0713 | No | ||
| 158 | YBX2 | 21400 | -4.595 | -0.0629 | No | ||
| 159 | SP3 | 21466 | -4.768 | -0.0553 | No | ||
| 160 | DNMT1 | 21530 | -4.995 | -0.0471 | No | ||
| 161 | USP52 | 21664 | -5.490 | -0.0410 | No | ||
| 162 | CDCA7 | 21685 | -5.595 | -0.0295 | No | ||
| 163 | MXD3 | 21708 | -5.712 | -0.0178 | No | ||
| 164 | MAZ | 21733 | -5.947 | -0.0057 | No | ||
| 165 | ERBB2IP | 21820 | -6.977 | 0.0058 | No |