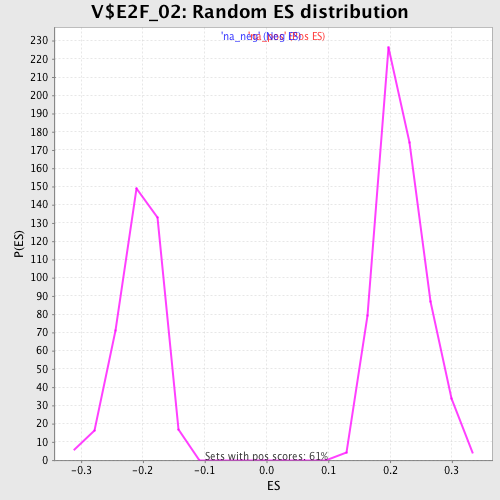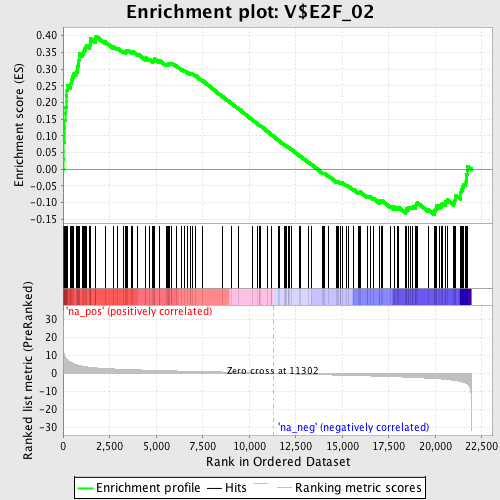
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set01_ATM_minus_versus_SCID |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_pos |
| GeneSet | V$E2F_02 |
| Enrichment Score (ES) | 0.39864683 |
| Normalized Enrichment Score (NES) | 1.8240391 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.061887383 |
| FWER p-Value | 0.106 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | SFRS10 | 19 | 13.401 | 0.0292 | Yes | ||
| 2 | NCL | 36 | 11.624 | 0.0545 | Yes | ||
| 3 | SYNCRIP | 38 | 11.556 | 0.0804 | Yes | ||
| 4 | EZH2 | 58 | 10.502 | 0.1031 | Yes | ||
| 5 | HNRPA2B1 | 59 | 10.466 | 0.1265 | Yes | ||
| 6 | MCM4 | 79 | 9.758 | 0.1476 | Yes | ||
| 7 | GPRC5B | 115 | 9.024 | 0.1662 | Yes | ||
| 8 | GSPT1 | 122 | 8.818 | 0.1857 | Yes | ||
| 9 | NASP | 168 | 8.269 | 0.2022 | Yes | ||
| 10 | ING3 | 170 | 8.243 | 0.2206 | Yes | ||
| 11 | SFRS7 | 206 | 7.661 | 0.2362 | Yes | ||
| 12 | CDC5L | 227 | 7.452 | 0.2520 | Yes | ||
| 13 | PRPF4B | 420 | 6.057 | 0.2567 | Yes | ||
| 14 | PRPS1 | 441 | 5.930 | 0.2691 | Yes | ||
| 15 | MSH5 | 508 | 5.562 | 0.2786 | Yes | ||
| 16 | NOLC1 | 581 | 5.207 | 0.2869 | Yes | ||
| 17 | PB1 | 693 | 4.835 | 0.2927 | Yes | ||
| 18 | STAG1 | 770 | 4.621 | 0.2996 | Yes | ||
| 19 | MAP3K7 | 785 | 4.594 | 0.3092 | Yes | ||
| 20 | TOPBP1 | 811 | 4.527 | 0.3182 | Yes | ||
| 21 | SFRS1 | 843 | 4.448 | 0.3268 | Yes | ||
| 22 | HNRPR | 858 | 4.419 | 0.3361 | Yes | ||
| 23 | PRPS2 | 860 | 4.418 | 0.3459 | Yes | ||
| 24 | RPS6KA5 | 1025 | 4.034 | 0.3474 | Yes | ||
| 25 | E2F3 | 1108 | 3.880 | 0.3524 | Yes | ||
| 26 | IPO7 | 1127 | 3.845 | 0.3602 | Yes | ||
| 27 | SLCO3A1 | 1216 | 3.697 | 0.3644 | Yes | ||
| 28 | AP4M1 | 1247 | 3.641 | 0.3712 | Yes | ||
| 29 | POLD3 | 1430 | 3.399 | 0.3705 | Yes | ||
| 30 | STAG2 | 1457 | 3.358 | 0.3768 | Yes | ||
| 31 | RBL1 | 1464 | 3.353 | 0.3841 | Yes | ||
| 32 | PTMA | 1471 | 3.346 | 0.3913 | Yes | ||
| 33 | HMGA1 | 1732 | 3.086 | 0.3863 | Yes | ||
| 34 | MAPK6 | 1746 | 3.072 | 0.3926 | Yes | ||
| 35 | PPIG | 1764 | 3.057 | 0.3986 | Yes | ||
| 36 | HTF9C | 2271 | 2.662 | 0.3814 | No | ||
| 37 | PCNA | 2701 | 2.431 | 0.3672 | No | ||
| 38 | IL4I1 | 2918 | 2.334 | 0.3625 | No | ||
| 39 | RANBP1 | 3228 | 2.211 | 0.3532 | No | ||
| 40 | ARHGAP11A | 3353 | 2.160 | 0.3524 | No | ||
| 41 | HNRPD | 3388 | 2.146 | 0.3556 | No | ||
| 42 | UGCGL1 | 3475 | 2.113 | 0.3564 | No | ||
| 43 | GATA1 | 3662 | 2.039 | 0.3525 | No | ||
| 44 | AK2 | 3741 | 2.013 | 0.3534 | No | ||
| 45 | ASXL2 | 4020 | 1.918 | 0.3450 | No | ||
| 46 | POU4F1 | 4414 | 1.797 | 0.3309 | No | ||
| 47 | MSH2 | 4451 | 1.783 | 0.3333 | No | ||
| 48 | ARHGAP6 | 4616 | 1.725 | 0.3296 | No | ||
| 49 | MYC | 4818 | 1.663 | 0.3241 | No | ||
| 50 | ID3 | 4852 | 1.653 | 0.3263 | No | ||
| 51 | PODN | 4914 | 1.630 | 0.3272 | No | ||
| 52 | PLAGL1 | 4924 | 1.627 | 0.3304 | No | ||
| 53 | FBXO5 | 5171 | 1.551 | 0.3226 | No | ||
| 54 | MCM6 | 5172 | 1.550 | 0.3261 | No | ||
| 55 | PAX6 | 5532 | 1.451 | 0.3129 | No | ||
| 56 | PHF5A | 5598 | 1.435 | 0.3131 | No | ||
| 57 | E2F1 | 5666 | 1.419 | 0.3132 | No | ||
| 58 | PEG3 | 5684 | 1.413 | 0.3156 | No | ||
| 59 | ACBD6 | 5734 | 1.399 | 0.3165 | No | ||
| 60 | PHF15 | 5799 | 1.382 | 0.3166 | No | ||
| 61 | MCM8 | 5839 | 1.373 | 0.3179 | No | ||
| 62 | KBTBD7 | 6080 | 1.316 | 0.3099 | No | ||
| 63 | TMPO | 6375 | 1.245 | 0.2992 | No | ||
| 64 | MAPT | 6530 | 1.211 | 0.2948 | No | ||
| 65 | PPM1D | 6687 | 1.174 | 0.2903 | No | ||
| 66 | PIM1 | 6823 | 1.141 | 0.2866 | No | ||
| 67 | CSPG2 | 6866 | 1.130 | 0.2872 | No | ||
| 68 | DMD | 6960 | 1.109 | 0.2855 | No | ||
| 69 | HCN3 | 7116 | 1.070 | 0.2808 | No | ||
| 70 | MCM3 | 7488 | 0.988 | 0.2659 | No | ||
| 71 | MRPL40 | 8576 | 0.740 | 0.2177 | No | ||
| 72 | ATF5 | 9046 | 0.628 | 0.1976 | No | ||
| 73 | HOXC10 | 9411 | 0.544 | 0.1821 | No | ||
| 74 | RTBDN | 10150 | 0.338 | 0.1490 | No | ||
| 75 | UNG | 10452 | 0.255 | 0.1357 | No | ||
| 76 | RQCD1 | 10542 | 0.231 | 0.1322 | No | ||
| 77 | SNRPD1 | 10547 | 0.229 | 0.1325 | No | ||
| 78 | GLRA3 | 10586 | 0.216 | 0.1312 | No | ||
| 79 | TBX6 | 11008 | 0.086 | 0.1121 | No | ||
| 80 | RET | 11200 | 0.029 | 0.1034 | No | ||
| 81 | STMN1 | 11211 | 0.027 | 0.1030 | No | ||
| 82 | ZIM2 | 11553 | -0.085 | 0.0875 | No | ||
| 83 | HS6ST3 | 11600 | -0.096 | 0.0856 | No | ||
| 84 | DNAJC9 | 11901 | -0.186 | 0.0723 | No | ||
| 85 | ALDH6A1 | 11952 | -0.198 | 0.0704 | No | ||
| 86 | PCSK1 | 11955 | -0.199 | 0.0708 | No | ||
| 87 | FMO4 | 12029 | -0.219 | 0.0679 | No | ||
| 88 | PRKDC | 12100 | -0.245 | 0.0653 | No | ||
| 89 | CORT | 12105 | -0.247 | 0.0656 | No | ||
| 90 | MCM7 | 12175 | -0.266 | 0.0631 | No | ||
| 91 | EED | 12272 | -0.293 | 0.0593 | No | ||
| 92 | PAQR4 | 12691 | -0.419 | 0.0411 | No | ||
| 93 | HNRPUL1 | 12768 | -0.442 | 0.0386 | No | ||
| 94 | RRM2 | 13177 | -0.554 | 0.0211 | No | ||
| 95 | ADAMTS2 | 13355 | -0.606 | 0.0143 | No | ||
| 96 | CASP8AP2 | 13952 | -0.766 | -0.0113 | No | ||
| 97 | EPHB1 | 14001 | -0.779 | -0.0118 | No | ||
| 98 | ZCCHC8 | 14040 | -0.789 | -0.0117 | No | ||
| 99 | ZBTB4 | 14269 | -0.843 | -0.0203 | No | ||
| 100 | RPS20 | 14667 | -0.949 | -0.0364 | No | ||
| 101 | RASAL2 | 14720 | -0.962 | -0.0366 | No | ||
| 102 | ILF3 | 14779 | -0.980 | -0.0371 | No | ||
| 103 | EIF4A1 | 14880 | -1.009 | -0.0394 | No | ||
| 104 | TRIM39 | 14918 | -1.026 | -0.0388 | No | ||
| 105 | FANCG | 14990 | -1.045 | -0.0397 | No | ||
| 106 | CBX3 | 15206 | -1.103 | -0.0471 | No | ||
| 107 | ARID4A | 15331 | -1.135 | -0.0503 | No | ||
| 108 | FANCD2 | 15609 | -1.209 | -0.0603 | No | ||
| 109 | PHC1 | 15865 | -1.276 | -0.0691 | No | ||
| 110 | SYNGR4 | 15905 | -1.285 | -0.0680 | No | ||
| 111 | DCK | 15954 | -1.299 | -0.0673 | No | ||
| 112 | GRIA4 | 16358 | -1.406 | -0.0827 | No | ||
| 113 | GABRB3 | 16371 | -1.408 | -0.0801 | No | ||
| 114 | KCNS2 | 16494 | -1.440 | -0.0824 | No | ||
| 115 | FHOD1 | 16660 | -1.484 | -0.0867 | No | ||
| 116 | NR6A1 | 16980 | -1.579 | -0.0978 | No | ||
| 117 | SLC9A5 | 16988 | -1.582 | -0.0946 | No | ||
| 118 | FANCC | 17098 | -1.617 | -0.0959 | No | ||
| 119 | SOAT1 | 17153 | -1.633 | -0.0948 | No | ||
| 120 | MCM2 | 17573 | -1.777 | -0.1100 | No | ||
| 121 | POLE2 | 17811 | -1.853 | -0.1167 | No | ||
| 122 | EFNA5 | 17815 | -1.855 | -0.1127 | No | ||
| 123 | CCNT1 | 17975 | -1.912 | -0.1157 | No | ||
| 124 | NELL2 | 18047 | -1.941 | -0.1146 | No | ||
| 125 | PKMYT1 | 18405 | -2.081 | -0.1263 | No | ||
| 126 | CLSPN | 18429 | -2.090 | -0.1227 | No | ||
| 127 | CDC25A | 18430 | -2.090 | -0.1180 | No | ||
| 128 | BRMS1L | 18546 | -2.138 | -0.1185 | No | ||
| 129 | KCNA6 | 18564 | -2.143 | -0.1145 | No | ||
| 130 | SEMA6A | 18650 | -2.183 | -0.1135 | No | ||
| 131 | USP37 | 18762 | -2.228 | -0.1136 | No | ||
| 132 | POLD1 | 18799 | -2.249 | -0.1102 | No | ||
| 133 | DNAJC5G | 18934 | -2.306 | -0.1112 | No | ||
| 134 | MYH10 | 18956 | -2.317 | -0.1069 | No | ||
| 135 | FKBP5 | 18982 | -2.330 | -0.1029 | No | ||
| 136 | SUV39H1 | 19043 | -2.361 | -0.1003 | No | ||
| 137 | CDC6 | 19624 | -2.669 | -0.1209 | No | ||
| 138 | NUP62 | 19937 | -2.850 | -0.1289 | No | ||
| 139 | TYRO3 | 19968 | -2.870 | -0.1238 | No | ||
| 140 | NRK | 20000 | -2.898 | -0.1187 | No | ||
| 141 | ACO2 | 20054 | -2.927 | -0.1146 | No | ||
| 142 | GMNN | 20067 | -2.936 | -0.1086 | No | ||
| 143 | POLE4 | 20237 | -3.051 | -0.1095 | No | ||
| 144 | SUMO1 | 20323 | -3.118 | -0.1064 | No | ||
| 145 | ATAD2 | 20395 | -3.167 | -0.1026 | No | ||
| 146 | POLR2A | 20527 | -3.291 | -0.1012 | No | ||
| 147 | STK35 | 20560 | -3.313 | -0.0952 | No | ||
| 148 | H2AFV | 20631 | -3.383 | -0.0909 | No | ||
| 149 | AP1S1 | 20990 | -3.808 | -0.0987 | No | ||
| 150 | BRPF1 | 21050 | -3.892 | -0.0927 | No | ||
| 151 | HIRA | 21077 | -3.929 | -0.0851 | No | ||
| 152 | H2AFZ | 21106 | -3.980 | -0.0775 | No | ||
| 153 | RAVER1 | 21359 | -4.509 | -0.0789 | No | ||
| 154 | E2F7 | 21360 | -4.512 | -0.0688 | No | ||
| 155 | YBX2 | 21400 | -4.595 | -0.0603 | No | ||
| 156 | SP3 | 21466 | -4.768 | -0.0526 | No | ||
| 157 | DNMT1 | 21530 | -4.995 | -0.0443 | No | ||
| 158 | TLE3 | 21646 | -5.426 | -0.0374 | No | ||
| 159 | USP52 | 21664 | -5.490 | -0.0258 | No | ||
| 160 | CDCA7 | 21685 | -5.595 | -0.0142 | No | ||
| 161 | MXD3 | 21708 | -5.712 | -0.0024 | No | ||
| 162 | MAZ | 21733 | -5.947 | 0.0098 | No |