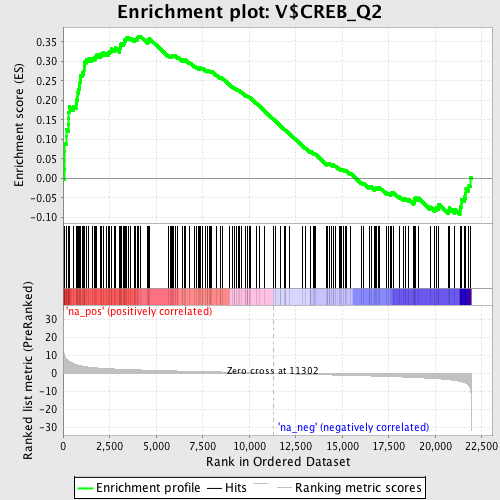
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set01_ATM_minus_versus_SCID |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_pos |
| GeneSet | V$CREB_Q2 |
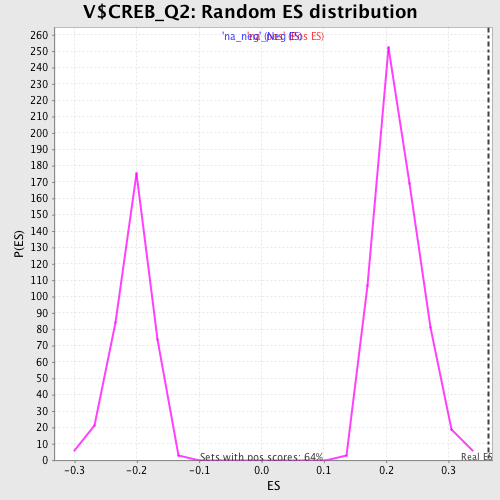
| Enrichment Score (ES) | 0.36467054 |
| Normalized Enrichment Score (NES) | 1.6720767 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.06552447 |
| FWER p-Value | 0.513 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | HNRPA2B1 | 59 | 10.466 | 0.0222 | Yes | ||
| 2 | DHX36 | 74 | 10.011 | 0.0455 | Yes | ||
| 3 | ETF1 | 77 | 9.911 | 0.0690 | Yes | ||
| 4 | DDX3X | 100 | 9.225 | 0.0900 | Yes | ||
| 5 | ING3 | 170 | 8.243 | 0.1064 | Yes | ||
| 6 | PABPC1 | 176 | 8.119 | 0.1256 | Yes | ||
| 7 | OSBPL9 | 273 | 7.038 | 0.1379 | Yes | ||
| 8 | ARIH1 | 286 | 6.974 | 0.1540 | Yes | ||
| 9 | MBNL2 | 310 | 6.749 | 0.1690 | Yes | ||
| 10 | NINJ1 | 333 | 6.588 | 0.1837 | Yes | ||
| 11 | RPS6KA3 | 571 | 5.239 | 0.1853 | Yes | ||
| 12 | SLC31A1 | 695 | 4.828 | 0.1912 | Yes | ||
| 13 | CLCN3 | 723 | 4.739 | 0.2012 | Yes | ||
| 14 | MAP3K7 | 785 | 4.594 | 0.2094 | Yes | ||
| 15 | SMS | 794 | 4.561 | 0.2199 | Yes | ||
| 16 | SFRS1 | 843 | 4.448 | 0.2283 | Yes | ||
| 17 | RAB6A | 862 | 4.413 | 0.2380 | Yes | ||
| 18 | BNIP3L | 904 | 4.310 | 0.2464 | Yes | ||
| 19 | ELOVL5 | 932 | 4.218 | 0.2552 | Yes | ||
| 20 | NDST1 | 951 | 4.190 | 0.2644 | Yes | ||
| 21 | PTP4A1 | 1028 | 4.027 | 0.2705 | Yes | ||
| 22 | ATP6V0C | 1121 | 3.858 | 0.2754 | Yes | ||
| 23 | NUP98 | 1125 | 3.851 | 0.2845 | Yes | ||
| 24 | UBQLN2 | 1133 | 3.831 | 0.2933 | Yes | ||
| 25 | GNAS | 1168 | 3.771 | 0.3007 | Yes | ||
| 26 | PDAP1 | 1278 | 3.598 | 0.3043 | Yes | ||
| 27 | EGR1 | 1388 | 3.442 | 0.3075 | Yes | ||
| 28 | CYLD | 1552 | 3.260 | 0.3078 | Yes | ||
| 29 | PLK4 | 1661 | 3.143 | 0.3103 | Yes | ||
| 30 | PPIG | 1764 | 3.057 | 0.3129 | Yes | ||
| 31 | IKBKB | 1816 | 3.007 | 0.3177 | Yes | ||
| 32 | WBP5 | 1992 | 2.855 | 0.3165 | Yes | ||
| 33 | OSR1 | 2071 | 2.789 | 0.3195 | Yes | ||
| 34 | PKP4 | 2161 | 2.728 | 0.3220 | Yes | ||
| 35 | ATF1 | 2314 | 2.637 | 0.3213 | Yes | ||
| 36 | TPM4 | 2435 | 2.568 | 0.3219 | Yes | ||
| 37 | SENP2 | 2498 | 2.538 | 0.3251 | Yes | ||
| 38 | EGR4 | 2573 | 2.496 | 0.3276 | Yes | ||
| 39 | NTRK2 | 2611 | 2.474 | 0.3318 | Yes | ||
| 40 | RAB7 | 2768 | 2.403 | 0.3304 | Yes | ||
| 41 | NDUFA10 | 2796 | 2.391 | 0.3348 | Yes | ||
| 42 | GTF2A1 | 3046 | 2.279 | 0.3288 | Yes | ||
| 43 | NR2E1 | 3053 | 2.277 | 0.3340 | Yes | ||
| 44 | CLSTN3 | 3079 | 2.267 | 0.3382 | Yes | ||
| 45 | AHI1 | 3088 | 2.263 | 0.3433 | Yes | ||
| 46 | RING1 | 3154 | 2.238 | 0.3456 | Yes | ||
| 47 | SPATA7 | 3267 | 2.191 | 0.3457 | Yes | ||
| 48 | CDC42 | 3285 | 2.187 | 0.3501 | Yes | ||
| 49 | SLC18A2 | 3296 | 2.183 | 0.3549 | Yes | ||
| 50 | ATF3 | 3370 | 2.151 | 0.3566 | Yes | ||
| 51 | CACNB2 | 3410 | 2.138 | 0.3599 | Yes | ||
| 52 | MRC2 | 3486 | 2.108 | 0.3615 | Yes | ||
| 53 | NR3C1 | 3635 | 2.049 | 0.3596 | Yes | ||
| 54 | ADAM9 | 3817 | 1.986 | 0.3560 | Yes | ||
| 55 | UCN | 3895 | 1.960 | 0.3572 | Yes | ||
| 56 | PRR3 | 3975 | 1.937 | 0.3581 | Yes | ||
| 57 | KIF17 | 4008 | 1.925 | 0.3613 | Yes | ||
| 58 | PHACTR3 | 4038 | 1.915 | 0.3645 | Yes | ||
| 59 | PQLC1 | 4133 | 1.885 | 0.3647 | Yes | ||
| 60 | GPM6B | 4530 | 1.761 | 0.3507 | No | ||
| 61 | WISP1 | 4575 | 1.743 | 0.3528 | No | ||
| 62 | FOSB | 4608 | 1.729 | 0.3555 | No | ||
| 63 | FGF6 | 4614 | 1.726 | 0.3593 | No | ||
| 64 | TRIB1 | 5655 | 1.422 | 0.3149 | No | ||
| 65 | RUSC1 | 5778 | 1.387 | 0.3126 | No | ||
| 66 | RELB | 5844 | 1.372 | 0.3129 | No | ||
| 67 | CENTA1 | 5877 | 1.362 | 0.3147 | No | ||
| 68 | CD2AP | 5915 | 1.352 | 0.3162 | No | ||
| 69 | AGPAT1 | 6032 | 1.327 | 0.3141 | No | ||
| 70 | CXCL16 | 6167 | 1.291 | 0.3110 | No | ||
| 71 | CRELD1 | 6411 | 1.237 | 0.3028 | No | ||
| 72 | GEM | 6426 | 1.234 | 0.3051 | No | ||
| 73 | PNRC1 | 6547 | 1.207 | 0.3024 | No | ||
| 74 | RBBP8 | 6555 | 1.206 | 0.3050 | No | ||
| 75 | ZMYND15 | 6770 | 1.155 | 0.2979 | No | ||
| 76 | LDHA | 7048 | 1.089 | 0.2878 | No | ||
| 77 | IRX4 | 7159 | 1.060 | 0.2852 | No | ||
| 78 | PITX2 | 7285 | 1.032 | 0.2820 | No | ||
| 79 | PPM2C | 7340 | 1.019 | 0.2819 | No | ||
| 80 | FBXL2 | 7358 | 1.016 | 0.2835 | No | ||
| 81 | KLF13 | 7470 | 0.991 | 0.2808 | No | ||
| 82 | CLDN7 | 7497 | 0.987 | 0.2820 | No | ||
| 83 | SCG2 | 7654 | 0.948 | 0.2771 | No | ||
| 84 | CX36 | 7750 | 0.931 | 0.2749 | No | ||
| 85 | RAB3A | 7774 | 0.925 | 0.2761 | No | ||
| 86 | SST | 7846 | 0.908 | 0.2750 | No | ||
| 87 | SMAD1 | 7927 | 0.889 | 0.2734 | No | ||
| 88 | CREM | 7942 | 0.886 | 0.2749 | No | ||
| 89 | SCAMP5 | 7996 | 0.872 | 0.2745 | No | ||
| 90 | EPHA2 | 8247 | 0.813 | 0.2650 | No | ||
| 91 | SMARCD1 | 8437 | 0.772 | 0.2581 | No | ||
| 92 | SYNGR3 | 8469 | 0.767 | 0.2585 | No | ||
| 93 | FOS | 8574 | 0.740 | 0.2555 | No | ||
| 94 | PER1 | 8937 | 0.652 | 0.2404 | No | ||
| 95 | COL5A3 | 9085 | 0.619 | 0.2352 | No | ||
| 96 | RAB25 | 9206 | 0.591 | 0.2311 | No | ||
| 97 | CSTF3 | 9224 | 0.588 | 0.2317 | No | ||
| 98 | TAGLN2 | 9342 | 0.561 | 0.2276 | No | ||
| 99 | HOXC10 | 9411 | 0.544 | 0.2258 | No | ||
| 100 | TGIF2 | 9467 | 0.533 | 0.2245 | No | ||
| 101 | FSTL5 | 9582 | 0.499 | 0.2205 | No | ||
| 102 | HHEX | 9811 | 0.435 | 0.2111 | No | ||
| 103 | DDX51 | 9824 | 0.430 | 0.2115 | No | ||
| 104 | HS3ST3A1 | 9882 | 0.415 | 0.2099 | No | ||
| 105 | FOXD3 | 9909 | 0.406 | 0.2097 | No | ||
| 106 | MAP1LC3A | 9925 | 0.403 | 0.2099 | No | ||
| 107 | IRX6 | 9998 | 0.382 | 0.2076 | No | ||
| 108 | CHGB | 10001 | 0.381 | 0.2084 | No | ||
| 109 | EVX1 | 10061 | 0.366 | 0.2065 | No | ||
| 110 | PTPRU | 10385 | 0.275 | 0.1923 | No | ||
| 111 | LMCD1 | 10541 | 0.231 | 0.1858 | No | ||
| 112 | PFAS | 10839 | 0.140 | 0.1725 | No | ||
| 113 | NEUROD6 | 11288 | 0.003 | 0.1519 | No | ||
| 114 | ID1 | 11419 | -0.040 | 0.1460 | No | ||
| 115 | SEMA4C | 11661 | -0.115 | 0.1352 | No | ||
| 116 | PNMA3 | 11694 | -0.125 | 0.1340 | No | ||
| 117 | DNAJC9 | 11901 | -0.186 | 0.1250 | No | ||
| 118 | ORMDL2 | 11949 | -0.198 | 0.1233 | No | ||
| 119 | PCSK1 | 11955 | -0.199 | 0.1236 | No | ||
| 120 | CALM2 | 12170 | -0.266 | 0.1144 | No | ||
| 121 | SREBF2 | 12843 | -0.462 | 0.0846 | No | ||
| 122 | DUSP1 | 13018 | -0.508 | 0.0778 | No | ||
| 123 | TEX14 | 13265 | -0.578 | 0.0679 | No | ||
| 124 | MYL6 | 13303 | -0.588 | 0.0676 | No | ||
| 125 | RNF44 | 13314 | -0.592 | 0.0685 | No | ||
| 126 | RFX5 | 13427 | -0.629 | 0.0649 | No | ||
| 127 | NUDT3 | 13493 | -0.647 | 0.0634 | No | ||
| 128 | DACT1 | 13548 | -0.662 | 0.0625 | No | ||
| 129 | WNT10A | 14165 | -0.821 | 0.0362 | No | ||
| 130 | RPL41 | 14180 | -0.824 | 0.0375 | No | ||
| 131 | MAFF | 14202 | -0.827 | 0.0385 | No | ||
| 132 | EGR3 | 14289 | -0.850 | 0.0366 | No | ||
| 133 | GADD45B | 14296 | -0.851 | 0.0383 | No | ||
| 134 | CTCF | 14413 | -0.882 | 0.0351 | No | ||
| 135 | PPP2R5B | 14506 | -0.909 | 0.0330 | No | ||
| 136 | SSTR2 | 14522 | -0.914 | 0.0345 | No | ||
| 137 | ADCY8 | 14635 | -0.939 | 0.0316 | No | ||
| 138 | RNF5 | 14832 | -0.994 | 0.0250 | No | ||
| 139 | TRIM39 | 14918 | -1.026 | 0.0235 | No | ||
| 140 | JAG1 | 14969 | -1.039 | 0.0237 | No | ||
| 141 | ERF | 15068 | -1.064 | 0.0217 | No | ||
| 142 | CDC14B | 15155 | -1.088 | 0.0204 | No | ||
| 143 | CBX3 | 15206 | -1.103 | 0.0207 | No | ||
| 144 | HHIP | 15416 | -1.157 | 0.0139 | No | ||
| 145 | MAF | 16023 | -1.316 | -0.0108 | No | ||
| 146 | MRRF | 16117 | -1.337 | -0.0119 | No | ||
| 147 | CRH | 16443 | -1.427 | -0.0235 | No | ||
| 148 | SNF1LK | 16473 | -1.434 | -0.0214 | No | ||
| 149 | FGF9 | 16548 | -1.454 | -0.0213 | No | ||
| 150 | NOL4 | 16745 | -1.511 | -0.0267 | No | ||
| 151 | HS3ST2 | 16788 | -1.521 | -0.0250 | No | ||
| 152 | GRM3 | 16820 | -1.532 | -0.0228 | No | ||
| 153 | LTBP1 | 16926 | -1.562 | -0.0239 | No | ||
| 154 | NR6A1 | 16980 | -1.579 | -0.0226 | No | ||
| 155 | BAT5 | 17386 | -1.715 | -0.0371 | No | ||
| 156 | NPTX1 | 17484 | -1.745 | -0.0374 | No | ||
| 157 | SDHB | 17604 | -1.785 | -0.0386 | No | ||
| 158 | WFDC3 | 17653 | -1.802 | -0.0365 | No | ||
| 159 | SLC38A1 | 17749 | -1.836 | -0.0365 | No | ||
| 160 | PPARGC1A | 18084 | -1.952 | -0.0472 | No | ||
| 161 | PAK3 | 18285 | -2.028 | -0.0515 | No | ||
| 162 | HERPUD1 | 18392 | -2.075 | -0.0515 | No | ||
| 163 | OGDH | 18565 | -2.143 | -0.0542 | No | ||
| 164 | ZFYVE27 | 18821 | -2.257 | -0.0606 | No | ||
| 165 | NR4A2 | 18880 | -2.284 | -0.0578 | No | ||
| 166 | MESDC1 | 18881 | -2.284 | -0.0524 | No | ||
| 167 | RAB24 | 18950 | -2.315 | -0.0500 | No | ||
| 168 | BRAF | 19077 | -2.378 | -0.0501 | No | ||
| 169 | DAAM2 | 19722 | -2.724 | -0.0732 | No | ||
| 170 | KCNA5 | 19981 | -2.878 | -0.0782 | No | ||
| 171 | GTF3C1 | 20066 | -2.936 | -0.0750 | No | ||
| 172 | RBMS2 | 20172 | -3.013 | -0.0727 | No | ||
| 173 | DSCR1 | 20193 | -3.023 | -0.0664 | No | ||
| 174 | GAK | 20716 | -3.463 | -0.0821 | No | ||
| 175 | NR4A3 | 20751 | -3.509 | -0.0753 | No | ||
| 176 | MAP3K13 | 21048 | -3.890 | -0.0797 | No | ||
| 177 | PLSCR3 | 21327 | -4.403 | -0.0819 | No | ||
| 178 | ZZEF1 | 21354 | -4.495 | -0.0724 | No | ||
| 179 | ELAVL1 | 21413 | -4.625 | -0.0641 | No | ||
| 180 | STAT3 | 21417 | -4.644 | -0.0531 | No | ||
| 181 | CAMK2D | 21559 | -5.093 | -0.0475 | No | ||
| 182 | PAFAH1B1 | 21628 | -5.352 | -0.0378 | No | ||
| 183 | TLE3 | 21646 | -5.426 | -0.0257 | No | ||
| 184 | NFATC1 | 21808 | -6.771 | -0.0169 | No | ||
| 185 | RAI1 | 21899 | -9.762 | 0.0022 | No |