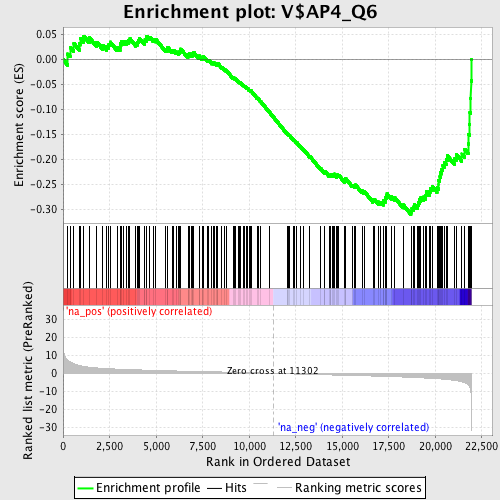
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set01_ATM_minus_versus_SCID |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_neg |
| GeneSet | V$AP4_Q6 |
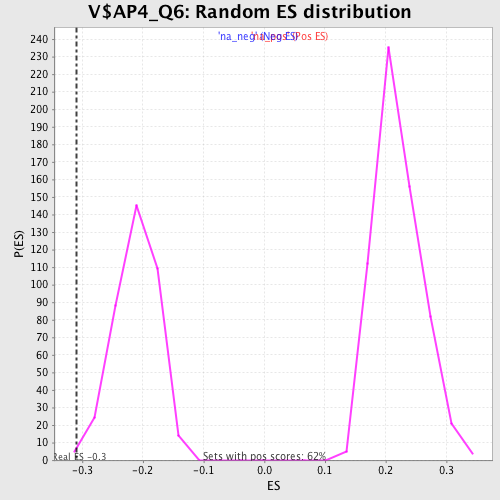
| Enrichment Score (ES) | -0.31001636 |
| Normalized Enrichment Score (NES) | -1.4645065 |
| Nominal p-value | 0.007792208 |
| FDR q-value | 0.4050493 |
| FWER p-Value | 0.968 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | TPM3 | 249 | 7.206 | 0.0111 | No | ||
| 2 | RAB27A | 378 | 6.311 | 0.0249 | No | ||
| 3 | RUNX1 | 580 | 5.210 | 0.0320 | No | ||
| 4 | SOST | 874 | 4.390 | 0.0323 | No | ||
| 5 | ARHGAP24 | 946 | 4.197 | 0.0421 | No | ||
| 6 | AKAP2 | 1111 | 3.878 | 0.0467 | No | ||
| 7 | TCF7 | 1405 | 3.424 | 0.0440 | No | ||
| 8 | ELA3A | 1807 | 3.019 | 0.0350 | No | ||
| 9 | GATA3 | 2129 | 2.751 | 0.0289 | No | ||
| 10 | WNT4 | 2355 | 2.610 | 0.0267 | No | ||
| 11 | FBXL11 | 2459 | 2.558 | 0.0300 | No | ||
| 12 | PALM2 | 2522 | 2.525 | 0.0350 | No | ||
| 13 | EYA1 | 2914 | 2.335 | 0.0244 | No | ||
| 14 | RASGEF1A | 3061 | 2.274 | 0.0248 | No | ||
| 15 | HDAC4 | 3069 | 2.271 | 0.0316 | No | ||
| 16 | PCDH12 | 3127 | 2.248 | 0.0360 | No | ||
| 17 | VAT1 | 3270 | 2.191 | 0.0363 | No | ||
| 18 | CACNB2 | 3410 | 2.138 | 0.0366 | No | ||
| 19 | BCL2L2 | 3519 | 2.096 | 0.0382 | No | ||
| 20 | SPOCK2 | 3591 | 2.066 | 0.0414 | No | ||
| 21 | PRKCQ | 3914 | 1.954 | 0.0327 | No | ||
| 22 | MYLK | 4002 | 1.928 | 0.0348 | No | ||
| 23 | NEUROG3 | 4041 | 1.914 | 0.0390 | No | ||
| 24 | ABHD4 | 4098 | 1.896 | 0.0424 | No | ||
| 25 | HFE2 | 4365 | 1.808 | 0.0358 | No | ||
| 26 | HSD11B1 | 4398 | 1.800 | 0.0400 | No | ||
| 27 | ZRANB1 | 4480 | 1.775 | 0.0418 | No | ||
| 28 | CADPS | 4493 | 1.772 | 0.0468 | No | ||
| 29 | LBX1 | 4663 | 1.710 | 0.0444 | No | ||
| 30 | ID3 | 4852 | 1.653 | 0.0409 | No | ||
| 31 | CPEB3 | 4982 | 1.611 | 0.0400 | No | ||
| 32 | PROK2 | 5527 | 1.453 | 0.0196 | No | ||
| 33 | SUV39H2 | 5592 | 1.435 | 0.0212 | No | ||
| 34 | ANXA8 | 5609 | 1.432 | 0.0249 | No | ||
| 35 | MID2 | 5867 | 1.366 | 0.0174 | No | ||
| 36 | RXRG | 5934 | 1.348 | 0.0186 | No | ||
| 37 | FILIP1 | 6097 | 1.313 | 0.0152 | No | ||
| 38 | GAS7 | 6217 | 1.280 | 0.0138 | No | ||
| 39 | NES | 6270 | 1.268 | 0.0154 | No | ||
| 40 | GRIN2B | 6287 | 1.264 | 0.0186 | No | ||
| 41 | LRRN5 | 6320 | 1.257 | 0.0210 | No | ||
| 42 | PDE3A | 6712 | 1.167 | 0.0067 | No | ||
| 43 | CHRNG | 6713 | 1.167 | 0.0104 | No | ||
| 44 | ASB16 | 6767 | 1.155 | 0.0116 | No | ||
| 45 | SOX12 | 6877 | 1.128 | 0.0101 | No | ||
| 46 | WNT9A | 6950 | 1.111 | 0.0103 | No | ||
| 47 | XPNPEP1 | 6968 | 1.107 | 0.0129 | No | ||
| 48 | HAPLN2 | 7023 | 1.093 | 0.0139 | No | ||
| 49 | PGF | 7302 | 1.028 | 0.0043 | No | ||
| 50 | PCYT1B | 7308 | 1.027 | 0.0073 | No | ||
| 51 | KLF13 | 7470 | 0.991 | 0.0030 | No | ||
| 52 | NPPA | 7504 | 0.985 | 0.0046 | No | ||
| 53 | LAT | 7545 | 0.978 | 0.0058 | No | ||
| 54 | RAB3A | 7774 | 0.925 | -0.0018 | No | ||
| 55 | KCNE1L | 7825 | 0.912 | -0.0012 | No | ||
| 56 | SCAMP5 | 7996 | 0.872 | -0.0063 | No | ||
| 57 | STOML2 | 8061 | 0.855 | -0.0066 | No | ||
| 58 | TNNT2 | 8125 | 0.841 | -0.0068 | No | ||
| 59 | EPHA2 | 8247 | 0.813 | -0.0098 | No | ||
| 60 | GNB3 | 8275 | 0.808 | -0.0085 | No | ||
| 61 | FGF8 | 8318 | 0.800 | -0.0080 | No | ||
| 62 | INHA | 8524 | 0.755 | -0.0150 | No | ||
| 63 | HOXB5 | 8681 | 0.718 | -0.0199 | No | ||
| 64 | GNG3 | 8784 | 0.691 | -0.0225 | No | ||
| 65 | RAB30 | 9131 | 0.607 | -0.0364 | No | ||
| 66 | ZNF385 | 9184 | 0.597 | -0.0370 | No | ||
| 67 | STARD13 | 9248 | 0.582 | -0.0380 | No | ||
| 68 | AICDA | 9429 | 0.542 | -0.0446 | No | ||
| 69 | GRIK3 | 9485 | 0.527 | -0.0455 | No | ||
| 70 | DHRS3 | 9518 | 0.517 | -0.0453 | No | ||
| 71 | FRAS1 | 9686 | 0.467 | -0.0515 | No | ||
| 72 | OBSCN | 9753 | 0.454 | -0.0531 | No | ||
| 73 | RASL12 | 9865 | 0.418 | -0.0569 | No | ||
| 74 | FOXD3 | 9909 | 0.406 | -0.0576 | No | ||
| 75 | EMILIN3 | 10009 | 0.379 | -0.0610 | No | ||
| 76 | COL7A1 | 10056 | 0.367 | -0.0620 | No | ||
| 77 | PTCH2 | 10097 | 0.355 | -0.0627 | No | ||
| 78 | HMGN2 | 10426 | 0.262 | -0.0769 | No | ||
| 79 | DLL3 | 10475 | 0.248 | -0.0783 | No | ||
| 80 | CACNA1G | 10621 | 0.207 | -0.0844 | No | ||
| 81 | CREB3L1 | 11070 | 0.071 | -0.1047 | No | ||
| 82 | PITX3 | 12034 | -0.221 | -0.1482 | No | ||
| 83 | ASB5 | 12056 | -0.229 | -0.1485 | No | ||
| 84 | NANOS1 | 12107 | -0.247 | -0.1500 | No | ||
| 85 | INPPL1 | 12157 | -0.262 | -0.1514 | No | ||
| 86 | MYBPH | 12378 | -0.325 | -0.1605 | No | ||
| 87 | SYTL2 | 12430 | -0.342 | -0.1618 | No | ||
| 88 | SMARCA2 | 12551 | -0.380 | -0.1661 | No | ||
| 89 | FBXO24 | 12742 | -0.434 | -0.1734 | No | ||
| 90 | EPOR | 12916 | -0.481 | -0.1799 | No | ||
| 91 | WNT2B | 13258 | -0.576 | -0.1937 | No | ||
| 92 | HOXC4 | 13823 | -0.729 | -0.2173 | No | ||
| 93 | TNFSF4 | 14048 | -0.791 | -0.2251 | No | ||
| 94 | ARID1A | 14066 | -0.796 | -0.2234 | No | ||
| 95 | MLLT7 | 14309 | -0.856 | -0.2319 | No | ||
| 96 | TNRC4 | 14348 | -0.866 | -0.2309 | No | ||
| 97 | PTK7 | 14392 | -0.875 | -0.2301 | No | ||
| 98 | ABLIM1 | 14473 | -0.899 | -0.2310 | No | ||
| 99 | RASL10B | 14502 | -0.908 | -0.2294 | No | ||
| 100 | FNDC5 | 14555 | -0.920 | -0.2290 | No | ||
| 101 | SIX5 | 14712 | -0.961 | -0.2331 | No | ||
| 102 | VGF | 14726 | -0.965 | -0.2307 | No | ||
| 103 | FMO5 | 14817 | -0.991 | -0.2317 | No | ||
| 104 | KCNN2 | 15126 | -1.081 | -0.2425 | No | ||
| 105 | RAMP2 | 15163 | -1.090 | -0.2407 | No | ||
| 106 | NXPH3 | 15182 | -1.097 | -0.2381 | No | ||
| 107 | TRPC4 | 15551 | -1.194 | -0.2513 | No | ||
| 108 | BACH1 | 15657 | -1.224 | -0.2523 | No | ||
| 109 | KCNQ4 | 15710 | -1.234 | -0.2508 | No | ||
| 110 | PABPC5 | 16061 | -1.325 | -0.2627 | No | ||
| 111 | FOSL2 | 16191 | -1.358 | -0.2644 | No | ||
| 112 | PLXNB3 | 16651 | -1.482 | -0.2808 | No | ||
| 113 | PHLDB1 | 16725 | -1.504 | -0.2795 | No | ||
| 114 | RGS7 | 16950 | -1.569 | -0.2849 | No | ||
| 115 | CNNM2 | 17065 | -1.607 | -0.2851 | No | ||
| 116 | FHL3 | 17200 | -1.648 | -0.2861 | No | ||
| 117 | TRAPPC3 | 17220 | -1.657 | -0.2818 | No | ||
| 118 | NDST2 | 17303 | -1.683 | -0.2803 | No | ||
| 119 | LOXL4 | 17325 | -1.691 | -0.2760 | No | ||
| 120 | EMP3 | 17364 | -1.707 | -0.2724 | No | ||
| 121 | EYA3 | 17402 | -1.720 | -0.2687 | No | ||
| 122 | ARHGEF2 | 17638 | -1.797 | -0.2739 | No | ||
| 123 | RASGRF2 | 17809 | -1.852 | -0.2759 | No | ||
| 124 | DOCK4 | 18272 | -2.024 | -0.2907 | No | ||
| 125 | ABTB2 | 18693 | -2.203 | -0.3031 | Yes | ||
| 126 | PTPRS | 18742 | -2.221 | -0.2984 | Yes | ||
| 127 | AMPH | 18822 | -2.257 | -0.2950 | Yes | ||
| 128 | MESDC1 | 18881 | -2.284 | -0.2905 | Yes | ||
| 129 | VPS45A | 19065 | -2.373 | -0.2915 | Yes | ||
| 130 | ELF4 | 19095 | -2.386 | -0.2853 | Yes | ||
| 131 | CBX6 | 19150 | -2.417 | -0.2803 | Yes | ||
| 132 | PRKACA | 19225 | -2.460 | -0.2760 | Yes | ||
| 133 | BSCL2 | 19377 | -2.534 | -0.2750 | Yes | ||
| 134 | ERN1 | 19490 | -2.602 | -0.2720 | Yes | ||
| 135 | FGF12 | 19506 | -2.610 | -0.2645 | Yes | ||
| 136 | TRAF4 | 19681 | -2.699 | -0.2641 | Yes | ||
| 137 | VAMP3 | 19721 | -2.724 | -0.2573 | Yes | ||
| 138 | TMOD4 | 19833 | -2.784 | -0.2537 | Yes | ||
| 139 | ARNT | 20092 | -2.961 | -0.2563 | Yes | ||
| 140 | TMOD3 | 20178 | -3.015 | -0.2508 | Yes | ||
| 141 | KCNMA1 | 20181 | -3.018 | -0.2415 | Yes | ||
| 142 | RBMS3 | 20247 | -3.056 | -0.2349 | Yes | ||
| 143 | SORBS1 | 20271 | -3.073 | -0.2263 | Yes | ||
| 144 | RTKN | 20332 | -3.122 | -0.2193 | Yes | ||
| 145 | RPS6KB1 | 20405 | -3.174 | -0.2127 | Yes | ||
| 146 | PICALM | 20475 | -3.247 | -0.2057 | Yes | ||
| 147 | ESRRA | 20585 | -3.344 | -0.2003 | Yes | ||
| 148 | WDTC1 | 20643 | -3.392 | -0.1923 | Yes | ||
| 149 | MAP3K13 | 21048 | -3.890 | -0.1987 | Yes | ||
| 150 | SYTL1 | 21120 | -4.007 | -0.1894 | Yes | ||
| 151 | UBTF | 21432 | -4.696 | -0.1890 | Yes | ||
| 152 | JMJD1C | 21579 | -5.171 | -0.1795 | Yes | ||
| 153 | GLTSCR1 | 21766 | -6.193 | -0.1687 | Yes | ||
| 154 | LCK | 21782 | -6.376 | -0.1495 | Yes | ||
| 155 | PRICKLE1 | 21831 | -7.167 | -0.1293 | Yes | ||
| 156 | BCL11B | 21858 | -7.702 | -0.1064 | Yes | ||
| 157 | CTDSP1 | 21906 | -10.094 | -0.0770 | Yes | ||
| 158 | ADRBK1 | 21916 | -11.172 | -0.0425 | Yes | ||
| 159 | RAI17 | 21938 | -14.036 | 0.0004 | Yes |