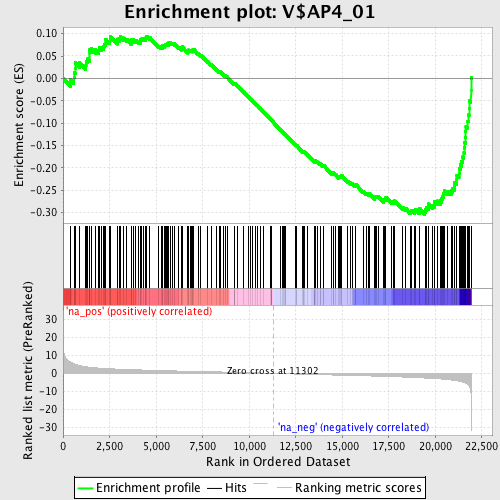
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set01_ATM_minus_versus_SCID |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_neg |
| GeneSet | V$AP4_01 |
| Enrichment Score (ES) | -0.30425033 |
| Normalized Enrichment Score (NES) | -1.4795189 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.41327778 |
| FWER p-Value | 0.939 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | TRPM7 | 394 | 6.197 | -0.0029 | No | ||
| 2 | RAP2C | 584 | 5.203 | 0.0012 | No | ||
| 3 | IGF1 | 591 | 5.167 | 0.0137 | No | ||
| 4 | FKBP3 | 652 | 4.935 | 0.0230 | No | ||
| 5 | NRD1 | 657 | 4.931 | 0.0350 | No | ||
| 6 | SOST | 874 | 4.390 | 0.0358 | No | ||
| 7 | CDK6 | 1221 | 3.690 | 0.0290 | No | ||
| 8 | PCBP2 | 1233 | 3.672 | 0.0375 | No | ||
| 9 | EPN2 | 1296 | 3.562 | 0.0434 | No | ||
| 10 | TCF7 | 1405 | 3.424 | 0.0469 | No | ||
| 11 | GSK3B | 1406 | 3.424 | 0.0553 | No | ||
| 12 | NAB2 | 1409 | 3.420 | 0.0636 | No | ||
| 13 | CUGBP1 | 1522 | 3.289 | 0.0666 | No | ||
| 14 | ARFIP2 | 1744 | 3.073 | 0.0640 | No | ||
| 15 | CDH13 | 1921 | 2.922 | 0.0630 | No | ||
| 16 | SPRY4 | 1952 | 2.886 | 0.0688 | No | ||
| 17 | POLR3E | 2079 | 2.786 | 0.0698 | No | ||
| 18 | MASP1 | 2186 | 2.710 | 0.0716 | No | ||
| 19 | SIN3A | 2221 | 2.686 | 0.0767 | No | ||
| 20 | DPF1 | 2283 | 2.655 | 0.0804 | No | ||
| 21 | FXC1 | 2285 | 2.654 | 0.0869 | No | ||
| 22 | PGGT1B | 2495 | 2.540 | 0.0835 | No | ||
| 23 | CACNG6 | 2520 | 2.527 | 0.0886 | No | ||
| 24 | PCTK1 | 2545 | 2.513 | 0.0937 | No | ||
| 25 | EYA1 | 2914 | 2.335 | 0.0825 | No | ||
| 26 | BCL11A | 2920 | 2.333 | 0.0880 | No | ||
| 27 | PJA2 | 3027 | 2.286 | 0.0888 | No | ||
| 28 | RASGEF1A | 3061 | 2.274 | 0.0928 | No | ||
| 29 | NPHP4 | 3235 | 2.207 | 0.0903 | No | ||
| 30 | CACNB2 | 3410 | 2.138 | 0.0876 | No | ||
| 31 | P2RY4 | 3654 | 2.041 | 0.0814 | No | ||
| 32 | GATA1 | 3662 | 2.039 | 0.0861 | No | ||
| 33 | FTS | 3772 | 2.000 | 0.0860 | No | ||
| 34 | OTOF | 3905 | 1.957 | 0.0848 | No | ||
| 35 | SGCG | 4058 | 1.907 | 0.0825 | No | ||
| 36 | CMKLR1 | 4139 | 1.883 | 0.0834 | No | ||
| 37 | GPC4 | 4165 | 1.875 | 0.0869 | No | ||
| 38 | KLF15 | 4235 | 1.853 | 0.0883 | No | ||
| 39 | FGF16 | 4327 | 1.821 | 0.0885 | No | ||
| 40 | POU4F1 | 4414 | 1.797 | 0.0890 | No | ||
| 41 | GRM7 | 4452 | 1.782 | 0.0917 | No | ||
| 42 | PCSK4 | 4488 | 1.773 | 0.0944 | No | ||
| 43 | LBX1 | 4663 | 1.710 | 0.0906 | No | ||
| 44 | NEUROD2 | 5147 | 1.559 | 0.0723 | No | ||
| 45 | CHD6 | 5283 | 1.522 | 0.0698 | No | ||
| 46 | SALL1 | 5345 | 1.505 | 0.0707 | No | ||
| 47 | MIS12 | 5356 | 1.502 | 0.0739 | No | ||
| 48 | UCP3 | 5449 | 1.473 | 0.0733 | No | ||
| 49 | GSH1 | 5475 | 1.466 | 0.0758 | No | ||
| 50 | MAPRE3 | 5535 | 1.451 | 0.0766 | No | ||
| 51 | RIT2 | 5594 | 1.435 | 0.0775 | No | ||
| 52 | LAG3 | 5675 | 1.414 | 0.0773 | No | ||
| 53 | CABP2 | 5687 | 1.412 | 0.0803 | No | ||
| 54 | RUSC1 | 5778 | 1.387 | 0.0795 | No | ||
| 55 | HES6 | 5878 | 1.362 | 0.0783 | No | ||
| 56 | BTBD11 | 5971 | 1.340 | 0.0774 | No | ||
| 57 | COL11A1 | 6213 | 1.281 | 0.0695 | No | ||
| 58 | NAV3 | 6349 | 1.250 | 0.0663 | No | ||
| 59 | SERPINE1 | 6362 | 1.248 | 0.0689 | No | ||
| 60 | ACCN5 | 6394 | 1.240 | 0.0705 | No | ||
| 61 | NXPH1 | 6698 | 1.171 | 0.0594 | No | ||
| 62 | PDE3A | 6712 | 1.167 | 0.0617 | No | ||
| 63 | ANGPTL2 | 6737 | 1.162 | 0.0635 | No | ||
| 64 | LIN28 | 6829 | 1.139 | 0.0621 | No | ||
| 65 | MOAP1 | 6886 | 1.125 | 0.0623 | No | ||
| 66 | DMD | 6960 | 1.109 | 0.0616 | No | ||
| 67 | SOX15 | 6977 | 1.106 | 0.0636 | No | ||
| 68 | HAPLN2 | 7023 | 1.093 | 0.0643 | No | ||
| 69 | PITX2 | 7285 | 1.032 | 0.0548 | No | ||
| 70 | MAPK8IP1 | 7407 | 1.004 | 0.0517 | No | ||
| 71 | POLD4 | 7773 | 0.926 | 0.0372 | No | ||
| 72 | USP2 | 7960 | 0.881 | 0.0308 | No | ||
| 73 | PSMD3 | 8256 | 0.811 | 0.0192 | No | ||
| 74 | SULT2B1 | 8410 | 0.780 | 0.0141 | No | ||
| 75 | HSD3B7 | 8442 | 0.772 | 0.0146 | No | ||
| 76 | PCBP4 | 8607 | 0.733 | 0.0089 | No | ||
| 77 | UNC13B | 8704 | 0.712 | 0.0062 | No | ||
| 78 | COL4A4 | 8808 | 0.685 | 0.0032 | No | ||
| 79 | ZNF385 | 9184 | 0.597 | -0.0126 | No | ||
| 80 | DOC2A | 9202 | 0.592 | -0.0119 | No | ||
| 81 | RAB25 | 9206 | 0.591 | -0.0106 | No | ||
| 82 | SNCA | 9379 | 0.552 | -0.0172 | No | ||
| 83 | RAPSN | 9685 | 0.468 | -0.0300 | No | ||
| 84 | ATP4A | 9945 | 0.397 | -0.0410 | No | ||
| 85 | EVX1 | 10061 | 0.366 | -0.0454 | No | ||
| 86 | SNAP25 | 10177 | 0.331 | -0.0498 | No | ||
| 87 | BAI2 | 10322 | 0.295 | -0.0557 | No | ||
| 88 | HMGN2 | 10426 | 0.262 | -0.0598 | No | ||
| 89 | TITF1 | 10608 | 0.210 | -0.0676 | No | ||
| 90 | EN1 | 10768 | 0.162 | -0.0745 | No | ||
| 91 | LLGL2 | 11166 | 0.039 | -0.0927 | No | ||
| 92 | GDPD2 | 11182 | 0.035 | -0.0933 | No | ||
| 93 | SMPX | 11201 | 0.029 | -0.0940 | No | ||
| 94 | GRIN2D | 11214 | 0.025 | -0.0945 | No | ||
| 95 | HRC | 11223 | 0.021 | -0.0948 | No | ||
| 96 | SEMA4C | 11661 | -0.115 | -0.1147 | No | ||
| 97 | GSC | 11814 | -0.162 | -0.1212 | No | ||
| 98 | KCNJ2 | 11817 | -0.162 | -0.1209 | No | ||
| 99 | RARA | 11911 | -0.188 | -0.1247 | No | ||
| 100 | PCSK1 | 11955 | -0.199 | -0.1262 | No | ||
| 101 | RHOG | 11958 | -0.200 | -0.1258 | No | ||
| 102 | PPARGC1B | 12459 | -0.351 | -0.1480 | No | ||
| 103 | HYAL2 | 12524 | -0.371 | -0.1500 | No | ||
| 104 | KCNE3 | 12553 | -0.380 | -0.1503 | No | ||
| 105 | HOXA11 | 12883 | -0.473 | -0.1643 | No | ||
| 106 | LMO1 | 12913 | -0.480 | -0.1645 | No | ||
| 107 | EPOR | 12916 | -0.481 | -0.1634 | No | ||
| 108 | NTN2L | 12966 | -0.493 | -0.1644 | No | ||
| 109 | SMARCA1 | 13129 | -0.539 | -0.1705 | No | ||
| 110 | NUDT3 | 13493 | -0.647 | -0.1856 | No | ||
| 111 | NR5A1 | 13508 | -0.652 | -0.1847 | No | ||
| 112 | PCDH1 | 13517 | -0.654 | -0.1834 | No | ||
| 113 | TRIM46 | 13543 | -0.661 | -0.1829 | No | ||
| 114 | KIF3C | 13664 | -0.688 | -0.1868 | No | ||
| 115 | SLC39A2 | 13814 | -0.727 | -0.1918 | No | ||
| 116 | HOXC4 | 13823 | -0.729 | -0.1904 | No | ||
| 117 | HOXA10 | 13989 | -0.775 | -0.1961 | No | ||
| 118 | KCNG4 | 13998 | -0.777 | -0.1945 | No | ||
| 119 | STARD10 | 14424 | -0.884 | -0.2119 | No | ||
| 120 | RASL10B | 14502 | -0.908 | -0.2132 | No | ||
| 121 | ACVR1C | 14503 | -0.908 | -0.2110 | No | ||
| 122 | RGS8 | 14616 | -0.936 | -0.2138 | No | ||
| 123 | FMO5 | 14817 | -0.991 | -0.2206 | No | ||
| 124 | ADCK4 | 14842 | -0.996 | -0.2192 | No | ||
| 125 | OAZ2 | 14903 | -1.017 | -0.2195 | No | ||
| 126 | NDUFB8 | 14945 | -1.035 | -0.2188 | No | ||
| 127 | CLPS | 14951 | -1.035 | -0.2165 | No | ||
| 128 | PPP1R9B | 15294 | -1.128 | -0.2295 | No | ||
| 129 | COL4A3 | 15448 | -1.167 | -0.2336 | No | ||
| 130 | LMO3 | 15539 | -1.191 | -0.2348 | No | ||
| 131 | SOX11 | 15692 | -1.231 | -0.2388 | No | ||
| 132 | WNT6 | 15730 | -1.240 | -0.2374 | No | ||
| 133 | DDIT3 | 16116 | -1.337 | -0.2519 | No | ||
| 134 | SESN2 | 16304 | -1.390 | -0.2570 | No | ||
| 135 | TMEM23 | 16412 | -1.419 | -0.2585 | No | ||
| 136 | H1F0 | 16483 | -1.436 | -0.2581 | No | ||
| 137 | DNAJB5 | 16756 | -1.514 | -0.2669 | No | ||
| 138 | SLC22A17 | 16764 | -1.516 | -0.2635 | No | ||
| 139 | PTPRT | 16844 | -1.538 | -0.2634 | No | ||
| 140 | LTBP1 | 16926 | -1.562 | -0.2633 | No | ||
| 141 | CSMD3 | 17238 | -1.661 | -0.2735 | No | ||
| 142 | ELL2 | 17249 | -1.664 | -0.2698 | No | ||
| 143 | LOXL4 | 17325 | -1.691 | -0.2691 | No | ||
| 144 | SEPT3 | 17345 | -1.700 | -0.2658 | No | ||
| 145 | KRTCAP2 | 17664 | -1.804 | -0.2760 | No | ||
| 146 | ANK2 | 17750 | -1.836 | -0.2754 | No | ||
| 147 | ITPKC | 17818 | -1.857 | -0.2739 | No | ||
| 148 | NFKBIL2 | 18244 | -2.011 | -0.2885 | No | ||
| 149 | VEGF | 18410 | -2.082 | -0.2910 | No | ||
| 150 | TLR7 | 18655 | -2.186 | -0.2968 | No | ||
| 151 | PTPRS | 18742 | -2.221 | -0.2953 | No | ||
| 152 | NR4A2 | 18880 | -2.284 | -0.2960 | No | ||
| 153 | NFIB | 18923 | -2.303 | -0.2923 | No | ||
| 154 | PLEC1 | 19137 | -2.411 | -0.2961 | No | ||
| 155 | OPCML | 19143 | -2.413 | -0.2904 | No | ||
| 156 | WDR23 | 19445 | -2.572 | -0.2979 | Yes | ||
| 157 | BAZ2A | 19481 | -2.598 | -0.2931 | Yes | ||
| 158 | SH3BP1 | 19524 | -2.620 | -0.2886 | Yes | ||
| 159 | ATP8A1 | 19619 | -2.667 | -0.2864 | Yes | ||
| 160 | SMAD7 | 19639 | -2.675 | -0.2807 | Yes | ||
| 161 | PFDN5 | 19863 | -2.805 | -0.2841 | Yes | ||
| 162 | NR1H2 | 19958 | -2.863 | -0.2813 | Yes | ||
| 163 | TYRO3 | 19968 | -2.870 | -0.2747 | Yes | ||
| 164 | CRB1 | 20114 | -2.974 | -0.2740 | Yes | ||
| 165 | SORBS1 | 20271 | -3.073 | -0.2737 | Yes | ||
| 166 | PTK2 | 20343 | -3.132 | -0.2692 | Yes | ||
| 167 | KCNK5 | 20396 | -3.167 | -0.2638 | Yes | ||
| 168 | LRP1 | 20444 | -3.214 | -0.2581 | Yes | ||
| 169 | EIF4E | 20471 | -3.243 | -0.2513 | Yes | ||
| 170 | KCNH2 | 20668 | -3.418 | -0.2519 | Yes | ||
| 171 | NUTF2 | 20850 | -3.630 | -0.2513 | Yes | ||
| 172 | BRUNOL4 | 20915 | -3.703 | -0.2451 | Yes | ||
| 173 | STARD3 | 21018 | -3.844 | -0.2404 | Yes | ||
| 174 | HIP2 | 21042 | -3.880 | -0.2319 | Yes | ||
| 175 | PPP2R5E | 21155 | -4.068 | -0.2270 | Yes | ||
| 176 | DLX1 | 21159 | -4.071 | -0.2172 | Yes | ||
| 177 | TCF8 | 21274 | -4.303 | -0.2118 | Yes | ||
| 178 | NDUFA4 | 21296 | -4.356 | -0.2021 | Yes | ||
| 179 | FOXP1 | 21341 | -4.453 | -0.1932 | Yes | ||
| 180 | STAT3 | 21417 | -4.644 | -0.1852 | Yes | ||
| 181 | MAP3K3 | 21465 | -4.765 | -0.1756 | Yes | ||
| 182 | SSBP2 | 21538 | -5.025 | -0.1666 | Yes | ||
| 183 | CLIC1 | 21553 | -5.073 | -0.1548 | Yes | ||
| 184 | JMJD1C | 21579 | -5.171 | -0.1432 | Yes | ||
| 185 | MTMR3 | 21621 | -5.316 | -0.1320 | Yes | ||
| 186 | SEMA7A | 21638 | -5.380 | -0.1195 | Yes | ||
| 187 | TLE3 | 21646 | -5.426 | -0.1065 | Yes | ||
| 188 | CTDSPL | 21748 | -6.080 | -0.0962 | Yes | ||
| 189 | GLTSCR1 | 21766 | -6.193 | -0.0818 | Yes | ||
| 190 | DNAJC14 | 21813 | -6.848 | -0.0670 | Yes | ||
| 191 | PRICKLE1 | 21831 | -7.167 | -0.0502 | Yes | ||
| 192 | ADRBK1 | 21916 | -11.172 | -0.0266 | Yes | ||
| 193 | PIK4CB | 21919 | -11.376 | 0.0013 | Yes |