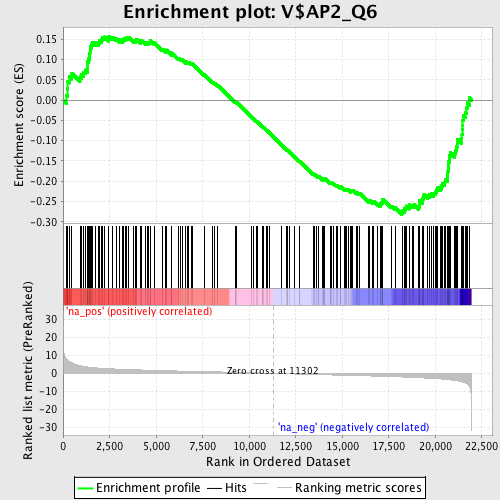
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set01_ATM_minus_versus_SCID |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_neg |
| GeneSet | V$AP2_Q6 |
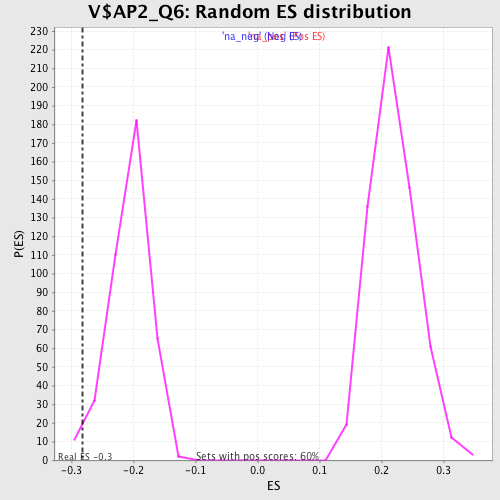
| Enrichment Score (ES) | -0.28195885 |
| Normalized Enrichment Score (NES) | -1.3713791 |
| Nominal p-value | 0.02238806 |
| FDR q-value | 0.31724072 |
| FWER p-Value | 0.999 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | PABPC1 | 176 | 8.119 | 0.0123 | No | ||
| 2 | CDK8 | 240 | 7.314 | 0.0277 | No | ||
| 3 | RAB10 | 252 | 7.194 | 0.0453 | No | ||
| 4 | DDX5 | 320 | 6.691 | 0.0589 | No | ||
| 5 | HDLBP | 474 | 5.763 | 0.0664 | No | ||
| 6 | VPS35 | 908 | 4.284 | 0.0572 | No | ||
| 7 | BCL3 | 989 | 4.105 | 0.0638 | No | ||
| 8 | E2F3 | 1108 | 3.880 | 0.0681 | No | ||
| 9 | CTNND2 | 1180 | 3.742 | 0.0743 | No | ||
| 10 | RRBP1 | 1299 | 3.560 | 0.0778 | No | ||
| 11 | SIRT1 | 1309 | 3.548 | 0.0863 | No | ||
| 12 | PDGFB | 1313 | 3.537 | 0.0950 | No | ||
| 13 | SPATA6 | 1356 | 3.478 | 0.1018 | No | ||
| 14 | CRK | 1421 | 3.408 | 0.1074 | No | ||
| 15 | POLD3 | 1430 | 3.399 | 0.1155 | No | ||
| 16 | POU2F1 | 1447 | 3.374 | 0.1233 | No | ||
| 17 | STAG2 | 1457 | 3.358 | 0.1313 | No | ||
| 18 | CUGBP1 | 1522 | 3.289 | 0.1366 | No | ||
| 19 | UBE2S | 1587 | 3.216 | 0.1417 | No | ||
| 20 | MAPK6 | 1746 | 3.072 | 0.1421 | No | ||
| 21 | ZZZ3 | 1908 | 2.935 | 0.1421 | No | ||
| 22 | WDR12 | 1950 | 2.887 | 0.1475 | No | ||
| 23 | ARL6IP2 | 2070 | 2.790 | 0.1490 | No | ||
| 24 | EBAG9 | 2117 | 2.759 | 0.1538 | No | ||
| 25 | CRBN | 2224 | 2.684 | 0.1557 | No | ||
| 26 | MAFB | 2443 | 2.564 | 0.1521 | No | ||
| 27 | FBXL11 | 2459 | 2.558 | 0.1578 | No | ||
| 28 | ARHGAP5 | 2666 | 2.449 | 0.1545 | No | ||
| 29 | CENPB | 2854 | 2.366 | 0.1518 | No | ||
| 30 | RAP1GDS1 | 3052 | 2.277 | 0.1485 | No | ||
| 31 | INSM1 | 3181 | 2.227 | 0.1482 | No | ||
| 32 | SDC1 | 3246 | 2.202 | 0.1508 | No | ||
| 33 | SUFU | 3336 | 2.166 | 0.1521 | No | ||
| 34 | RASA1 | 3405 | 2.139 | 0.1543 | No | ||
| 35 | EP300 | 3501 | 2.101 | 0.1552 | No | ||
| 36 | GATA2 | 3796 | 1.994 | 0.1467 | No | ||
| 37 | HPN | 3884 | 1.962 | 0.1477 | No | ||
| 38 | MYADM | 3944 | 1.946 | 0.1498 | No | ||
| 39 | GPC4 | 4165 | 1.875 | 0.1444 | No | ||
| 40 | FKBP14 | 4187 | 1.866 | 0.1481 | No | ||
| 41 | RASIP1 | 4429 | 1.790 | 0.1416 | No | ||
| 42 | FST | 4509 | 1.766 | 0.1424 | No | ||
| 43 | ALG5 | 4603 | 1.730 | 0.1424 | No | ||
| 44 | NRG1 | 4674 | 1.708 | 0.1435 | No | ||
| 45 | ORC6L | 4707 | 1.697 | 0.1463 | No | ||
| 46 | IGFBP7 | 4887 | 1.641 | 0.1422 | No | ||
| 47 | SALL1 | 5345 | 1.505 | 0.1250 | No | ||
| 48 | TEAD2 | 5526 | 1.454 | 0.1203 | No | ||
| 49 | MAPRE3 | 5535 | 1.451 | 0.1236 | No | ||
| 50 | GATA6 | 5819 | 1.378 | 0.1141 | No | ||
| 51 | RELB | 5844 | 1.372 | 0.1164 | No | ||
| 52 | GAS7 | 6217 | 1.280 | 0.1025 | No | ||
| 53 | WNT7A | 6322 | 1.256 | 0.1009 | No | ||
| 54 | IGSF4 | 6412 | 1.237 | 0.0999 | No | ||
| 55 | SPRY2 | 6601 | 1.196 | 0.0943 | No | ||
| 56 | PPM1E | 6679 | 1.176 | 0.0937 | No | ||
| 57 | ANGPTL2 | 6737 | 1.162 | 0.0940 | No | ||
| 58 | SOX12 | 6877 | 1.128 | 0.0904 | No | ||
| 59 | ZDHHC2 | 6933 | 1.113 | 0.0907 | No | ||
| 60 | LHX1 | 7603 | 0.961 | 0.0624 | No | ||
| 61 | CRLF1 | 8038 | 0.861 | 0.0446 | No | ||
| 62 | KCNH5 | 8146 | 0.835 | 0.0418 | No | ||
| 63 | FAF1 | 8302 | 0.803 | 0.0367 | No | ||
| 64 | C1ORF24 | 9249 | 0.581 | -0.0053 | No | ||
| 65 | SFRP2 | 9280 | 0.574 | -0.0053 | No | ||
| 66 | PDE5A | 9327 | 0.564 | -0.0060 | No | ||
| 67 | WNT1 | 10135 | 0.345 | -0.0422 | No | ||
| 68 | GPR3 | 10203 | 0.326 | -0.0445 | No | ||
| 69 | APBA1 | 10390 | 0.274 | -0.0523 | No | ||
| 70 | HMGN2 | 10426 | 0.262 | -0.0533 | No | ||
| 71 | SLC30A5 | 10688 | 0.189 | -0.0648 | No | ||
| 72 | FBXO11 | 10735 | 0.177 | -0.0664 | No | ||
| 73 | HCST | 10750 | 0.171 | -0.0667 | No | ||
| 74 | SOX2 | 10930 | 0.108 | -0.0746 | No | ||
| 75 | MBD3 | 10974 | 0.097 | -0.0763 | No | ||
| 76 | CREB3L1 | 11070 | 0.071 | -0.0805 | No | ||
| 77 | SMOX | 11718 | -0.133 | -0.1099 | No | ||
| 78 | ARPC1A | 12005 | -0.212 | -0.1225 | No | ||
| 79 | PITX3 | 12034 | -0.221 | -0.1233 | No | ||
| 80 | MNT | 12040 | -0.223 | -0.1229 | No | ||
| 81 | VEGFB | 12174 | -0.266 | -0.1284 | No | ||
| 82 | WNT3 | 12450 | -0.349 | -0.1401 | No | ||
| 83 | GLTP | 12692 | -0.419 | -0.1501 | No | ||
| 84 | FGF11 | 12725 | -0.429 | -0.1505 | No | ||
| 85 | BANP | 13442 | -0.632 | -0.1818 | No | ||
| 86 | MTA2 | 13520 | -0.656 | -0.1837 | No | ||
| 87 | KLC2 | 13633 | -0.680 | -0.1872 | No | ||
| 88 | FZD5 | 13726 | -0.706 | -0.1896 | No | ||
| 89 | CNNM1 | 13729 | -0.706 | -0.1880 | No | ||
| 90 | ICA1 | 13931 | -0.760 | -0.1953 | No | ||
| 91 | CNNM4 | 13976 | -0.772 | -0.1954 | No | ||
| 92 | EPHB1 | 14001 | -0.779 | -0.1945 | No | ||
| 93 | SP4 | 14045 | -0.791 | -0.1945 | No | ||
| 94 | ARID1A | 14066 | -0.796 | -0.1934 | No | ||
| 95 | LMX1A | 14354 | -0.867 | -0.2044 | No | ||
| 96 | HCN1 | 14385 | -0.874 | -0.2036 | No | ||
| 97 | CTCF | 14413 | -0.882 | -0.2027 | No | ||
| 98 | RCOR2 | 14551 | -0.919 | -0.2067 | No | ||
| 99 | PPP6C | 14680 | -0.954 | -0.2101 | No | ||
| 100 | RASAL2 | 14720 | -0.962 | -0.2095 | No | ||
| 101 | TERF2 | 14887 | -1.011 | -0.2146 | No | ||
| 102 | TRIM39 | 14918 | -1.026 | -0.2134 | No | ||
| 103 | KCNN2 | 15126 | -1.081 | -0.2202 | No | ||
| 104 | NXPH3 | 15182 | -1.097 | -0.2200 | No | ||
| 105 | CNP | 15242 | -1.113 | -0.2199 | No | ||
| 106 | NOTCH3 | 15321 | -1.133 | -0.2207 | No | ||
| 107 | CPEB4 | 15466 | -1.173 | -0.2243 | No | ||
| 108 | SLC7A10 | 15478 | -1.175 | -0.2219 | No | ||
| 109 | GALNT14 | 15538 | -1.191 | -0.2216 | No | ||
| 110 | APLP1 | 15754 | -1.245 | -0.2284 | No | ||
| 111 | BSN | 15818 | -1.265 | -0.2281 | No | ||
| 112 | PSCD2 | 15917 | -1.288 | -0.2294 | No | ||
| 113 | TRIM28 | 16418 | -1.420 | -0.2488 | No | ||
| 114 | H1F0 | 16483 | -1.436 | -0.2481 | No | ||
| 115 | GALNT10 | 16615 | -1.472 | -0.2504 | No | ||
| 116 | GUCY1A2 | 16664 | -1.485 | -0.2489 | No | ||
| 117 | SPTBN4 | 16918 | -1.559 | -0.2566 | No | ||
| 118 | ATF7IP | 17055 | -1.602 | -0.2589 | No | ||
| 119 | CNNM2 | 17065 | -1.607 | -0.2552 | No | ||
| 120 | DUSP15 | 17099 | -1.618 | -0.2527 | No | ||
| 121 | HOXB2 | 17142 | -1.630 | -0.2505 | No | ||
| 122 | ATP5G2 | 17156 | -1.634 | -0.2470 | No | ||
| 123 | USP48 | 17178 | -1.641 | -0.2439 | No | ||
| 124 | WFDC3 | 17653 | -1.802 | -0.2612 | No | ||
| 125 | PBX3 | 17849 | -1.869 | -0.2654 | No | ||
| 126 | PLXNC1 | 18210 | -2.000 | -0.2769 | Yes | ||
| 127 | MMD | 18236 | -2.008 | -0.2731 | Yes | ||
| 128 | REV3L | 18340 | -2.055 | -0.2726 | Yes | ||
| 129 | RAB3IP | 18344 | -2.056 | -0.2676 | Yes | ||
| 130 | SLC25A12 | 18422 | -2.087 | -0.2659 | Yes | ||
| 131 | CDC25A | 18430 | -2.090 | -0.2610 | Yes | ||
| 132 | EFEMP2 | 18593 | -2.152 | -0.2630 | Yes | ||
| 133 | GRK5 | 18611 | -2.162 | -0.2584 | Yes | ||
| 134 | PPFIA1 | 18771 | -2.231 | -0.2601 | Yes | ||
| 135 | THRAP1 | 18853 | -2.271 | -0.2581 | Yes | ||
| 136 | BAHD1 | 19118 | -2.400 | -0.2643 | Yes | ||
| 137 | SHB | 19128 | -2.407 | -0.2586 | Yes | ||
| 138 | RAB35 | 19147 | -2.417 | -0.2534 | Yes | ||
| 139 | CBX6 | 19150 | -2.417 | -0.2474 | Yes | ||
| 140 | ACTR1A | 19311 | -2.504 | -0.2485 | Yes | ||
| 141 | DNMT3A | 19318 | -2.507 | -0.2425 | Yes | ||
| 142 | CAMTA2 | 19352 | -2.520 | -0.2377 | Yes | ||
| 143 | SLC25A28 | 19390 | -2.542 | -0.2330 | Yes | ||
| 144 | PLAGL2 | 19597 | -2.657 | -0.2358 | Yes | ||
| 145 | PLCB3 | 19683 | -2.699 | -0.2330 | Yes | ||
| 146 | SFXN2 | 19776 | -2.753 | -0.2303 | Yes | ||
| 147 | COL11A2 | 19914 | -2.838 | -0.2295 | Yes | ||
| 148 | LDB1 | 20007 | -2.902 | -0.2264 | Yes | ||
| 149 | RAB22A | 20059 | -2.931 | -0.2214 | Yes | ||
| 150 | CREB3 | 20115 | -2.975 | -0.2165 | Yes | ||
| 151 | RASGRP2 | 20268 | -3.070 | -0.2157 | Yes | ||
| 152 | DNTTIP1 | 20346 | -3.133 | -0.2114 | Yes | ||
| 153 | ACTB | 20378 | -3.156 | -0.2049 | Yes | ||
| 154 | UBE2B | 20517 | -3.289 | -0.2030 | Yes | ||
| 155 | IRF1 | 20521 | -3.289 | -0.1949 | Yes | ||
| 156 | CDK9 | 20639 | -3.389 | -0.1918 | Yes | ||
| 157 | MAST2 | 20654 | -3.411 | -0.1839 | Yes | ||
| 158 | HS3ST3B1 | 20678 | -3.426 | -0.1764 | Yes | ||
| 159 | MYST2 | 20696 | -3.444 | -0.1685 | Yes | ||
| 160 | EXOSC8 | 20713 | -3.458 | -0.1606 | Yes | ||
| 161 | CAMK4 | 20719 | -3.468 | -0.1521 | Yes | ||
| 162 | CRSP7 | 20747 | -3.506 | -0.1445 | Yes | ||
| 163 | POFUT1 | 20778 | -3.537 | -0.1371 | Yes | ||
| 164 | ATP5H | 20804 | -3.572 | -0.1292 | Yes | ||
| 165 | BRPF1 | 21050 | -3.892 | -0.1307 | Yes | ||
| 166 | SMARCD2 | 21099 | -3.965 | -0.1230 | Yes | ||
| 167 | PPP2R5E | 21155 | -4.068 | -0.1153 | Yes | ||
| 168 | FMNL1 | 21168 | -4.083 | -0.1056 | Yes | ||
| 169 | WAC | 21182 | -4.103 | -0.0960 | Yes | ||
| 170 | ZDHHC14 | 21422 | -4.658 | -0.0953 | Yes | ||
| 171 | UBTF | 21432 | -4.696 | -0.0839 | Yes | ||
| 172 | NRF1 | 21445 | -4.729 | -0.0726 | Yes | ||
| 173 | SP3 | 21466 | -4.768 | -0.0616 | Yes | ||
| 174 | BCL2 | 21470 | -4.794 | -0.0497 | Yes | ||
| 175 | LHX2 | 21501 | -4.887 | -0.0388 | Yes | ||
| 176 | PHF12 | 21607 | -5.257 | -0.0304 | Yes | ||
| 177 | ANKRD27 | 21656 | -5.464 | -0.0189 | Yes | ||
| 178 | TLN1 | 21707 | -5.706 | -0.0069 | Yes | ||
| 179 | MAPRE1 | 21828 | -7.143 | 0.0055 | Yes |