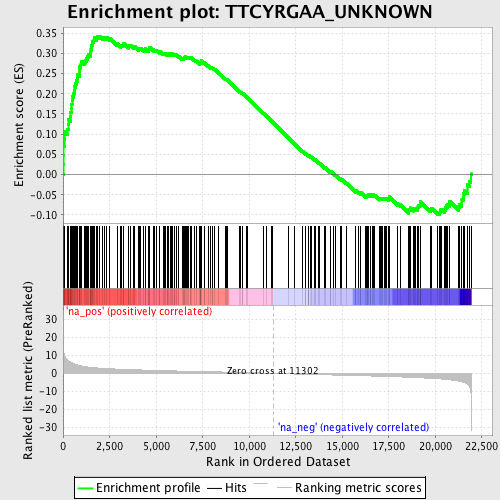
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set01_ATM_minus_versus_SCID |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_pos |
| GeneSet | TTCYRGAA_UNKNOWN |
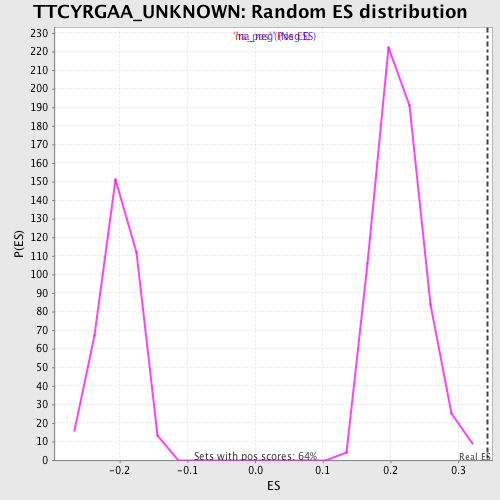
| Enrichment Score (ES) | 0.34265655 |
| Normalized Enrichment Score (NES) | 1.6010273 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.092823915 |
| FWER p-Value | 0.78 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | SFRS10 | 19 | 13.401 | 0.0247 | Yes | ||
| 2 | MTPN | 29 | 12.372 | 0.0478 | Yes | ||
| 3 | SYNCRIP | 38 | 11.556 | 0.0695 | Yes | ||
| 4 | HNRPA2B1 | 59 | 10.466 | 0.0885 | Yes | ||
| 5 | DNAJA1 | 83 | 9.638 | 0.1058 | Yes | ||
| 6 | PPP1R7 | 244 | 7.261 | 0.1123 | Yes | ||
| 7 | TPP2 | 288 | 6.967 | 0.1236 | Yes | ||
| 8 | CYP39A1 | 298 | 6.839 | 0.1362 | Yes | ||
| 9 | PAFAH1B2 | 385 | 6.246 | 0.1441 | Yes | ||
| 10 | TXNDC9 | 410 | 6.125 | 0.1547 | Yes | ||
| 11 | PHF6 | 456 | 5.849 | 0.1637 | Yes | ||
| 12 | HSPD1 | 465 | 5.801 | 0.1744 | Yes | ||
| 13 | XPO1 | 482 | 5.700 | 0.1846 | Yes | ||
| 14 | NUP153 | 524 | 5.472 | 0.1931 | Yes | ||
| 15 | RPS6KA3 | 571 | 5.239 | 0.2010 | Yes | ||
| 16 | YWHAG | 608 | 5.096 | 0.2090 | Yes | ||
| 17 | CDH22 | 634 | 4.993 | 0.2174 | Yes | ||
| 18 | ELP4 | 681 | 4.866 | 0.2245 | Yes | ||
| 19 | RAB18 | 713 | 4.768 | 0.2322 | Yes | ||
| 20 | CCT4 | 750 | 4.659 | 0.2394 | Yes | ||
| 21 | CNIH | 782 | 4.600 | 0.2467 | Yes | ||
| 22 | PIK3R1 | 880 | 4.360 | 0.2506 | Yes | ||
| 23 | HSPB1 | 886 | 4.353 | 0.2586 | Yes | ||
| 24 | JARID2 | 891 | 4.340 | 0.2667 | Yes | ||
| 25 | STAMBP | 959 | 4.175 | 0.2716 | Yes | ||
| 26 | YWHAQ | 965 | 4.155 | 0.2793 | Yes | ||
| 27 | NUP98 | 1125 | 3.851 | 0.2793 | Yes | ||
| 28 | ST13 | 1199 | 3.718 | 0.2830 | Yes | ||
| 29 | TGIF | 1266 | 3.612 | 0.2869 | Yes | ||
| 30 | PDGFB | 1313 | 3.537 | 0.2915 | Yes | ||
| 31 | MORF4L2 | 1347 | 3.499 | 0.2967 | Yes | ||
| 32 | STAG2 | 1457 | 3.358 | 0.2980 | Yes | ||
| 33 | TOP1 | 1486 | 3.326 | 0.3031 | Yes | ||
| 34 | PTHLH | 1496 | 3.318 | 0.3090 | Yes | ||
| 35 | CCT8 | 1520 | 3.291 | 0.3142 | Yes | ||
| 36 | CHORDC1 | 1536 | 3.276 | 0.3198 | Yes | ||
| 37 | GPATC2 | 1562 | 3.249 | 0.3248 | Yes | ||
| 38 | ADRA2C | 1593 | 3.209 | 0.3295 | Yes | ||
| 39 | ATP6V0D2 | 1656 | 3.149 | 0.3327 | Yes | ||
| 40 | AOAH | 1662 | 3.143 | 0.3384 | Yes | ||
| 41 | SOD2 | 1804 | 3.024 | 0.3377 | Yes | ||
| 42 | DNAJB4 | 1839 | 2.990 | 0.3418 | Yes | ||
| 43 | PRDX6 | 1942 | 2.895 | 0.3427 | Yes | ||
| 44 | EGR2 | 2110 | 2.763 | 0.3402 | No | ||
| 45 | EIF5B | 2248 | 2.672 | 0.3390 | No | ||
| 46 | ATP2C1 | 2352 | 2.614 | 0.3393 | No | ||
| 47 | HOXA3 | 2496 | 2.540 | 0.3375 | No | ||
| 48 | MGAT3 | 2896 | 2.343 | 0.3236 | No | ||
| 49 | MAP3K6 | 3076 | 2.268 | 0.3197 | No | ||
| 50 | MAT2A | 3156 | 2.238 | 0.3203 | No | ||
| 51 | CEPT1 | 3221 | 2.214 | 0.3216 | No | ||
| 52 | CLU | 3264 | 2.192 | 0.3239 | No | ||
| 53 | EP300 | 3501 | 2.101 | 0.3170 | No | ||
| 54 | HSPB2 | 3525 | 2.092 | 0.3199 | No | ||
| 55 | SPIN | 3617 | 2.056 | 0.3197 | No | ||
| 56 | HAT1 | 3804 | 1.990 | 0.3149 | No | ||
| 57 | CRYAB | 3824 | 1.983 | 0.3178 | No | ||
| 58 | TWIST2 | 4065 | 1.905 | 0.3104 | No | ||
| 59 | DAZL | 4088 | 1.898 | 0.3130 | No | ||
| 60 | GPC4 | 4165 | 1.875 | 0.3131 | No | ||
| 61 | FGF16 | 4327 | 1.821 | 0.3091 | No | ||
| 62 | EVC2 | 4411 | 1.798 | 0.3087 | No | ||
| 63 | ACVR1 | 4432 | 1.789 | 0.3112 | No | ||
| 64 | SERPINA10 | 4582 | 1.739 | 0.3077 | No | ||
| 65 | ARHGEF12 | 4612 | 1.727 | 0.3096 | No | ||
| 66 | ARHGAP6 | 4616 | 1.725 | 0.3128 | No | ||
| 67 | FBXO30 | 4630 | 1.720 | 0.3155 | No | ||
| 68 | UCHL1 | 4845 | 1.656 | 0.3088 | No | ||
| 69 | SERPINH1 | 4936 | 1.625 | 0.3077 | No | ||
| 70 | NUDC | 5010 | 1.602 | 0.3074 | No | ||
| 71 | FBXO5 | 5171 | 1.551 | 0.3030 | No | ||
| 72 | RAB2B | 5202 | 1.541 | 0.3046 | No | ||
| 73 | FBXO33 | 5391 | 1.492 | 0.2988 | No | ||
| 74 | SEMA3A | 5424 | 1.484 | 0.3001 | No | ||
| 75 | RTN1 | 5510 | 1.457 | 0.2990 | No | ||
| 76 | LGALS8 | 5597 | 1.435 | 0.2978 | No | ||
| 77 | PTDSR | 5603 | 1.434 | 0.3003 | No | ||
| 78 | E2F1 | 5666 | 1.419 | 0.3001 | No | ||
| 79 | SIX3 | 5791 | 1.384 | 0.2971 | No | ||
| 80 | PHF15 | 5799 | 1.382 | 0.2994 | No | ||
| 81 | PGM1 | 5855 | 1.368 | 0.2994 | No | ||
| 82 | BTBD11 | 5971 | 1.340 | 0.2967 | No | ||
| 83 | STIP1 | 6007 | 1.332 | 0.2976 | No | ||
| 84 | AGTR2 | 6094 | 1.314 | 0.2962 | No | ||
| 85 | GAS7 | 6217 | 1.280 | 0.2930 | No | ||
| 86 | LRRC17 | 6388 | 1.242 | 0.2875 | No | ||
| 87 | FRAG1 | 6470 | 1.224 | 0.2862 | No | ||
| 88 | FLNC | 6494 | 1.218 | 0.2874 | No | ||
| 89 | PRDM2 | 6520 | 1.213 | 0.2886 | No | ||
| 90 | MAPT | 6530 | 1.211 | 0.2905 | No | ||
| 91 | SATB2 | 6562 | 1.205 | 0.2913 | No | ||
| 92 | CDKN1A | 6632 | 1.188 | 0.2904 | No | ||
| 93 | SELP | 6703 | 1.169 | 0.2894 | No | ||
| 94 | TNMD | 6761 | 1.156 | 0.2890 | No | ||
| 95 | CDK5R2 | 6824 | 1.140 | 0.2883 | No | ||
| 96 | SAG | 6863 | 1.131 | 0.2887 | No | ||
| 97 | CXXC4 | 6895 | 1.124 | 0.2895 | No | ||
| 98 | DMRTB1 | 7076 | 1.081 | 0.2832 | No | ||
| 99 | ELOVL1 | 7166 | 1.058 | 0.2812 | No | ||
| 100 | PPM2C | 7340 | 1.019 | 0.2751 | No | ||
| 101 | CYP46A1 | 7365 | 1.013 | 0.2760 | No | ||
| 102 | LMOD1 | 7388 | 1.007 | 0.2769 | No | ||
| 103 | PER2 | 7401 | 1.005 | 0.2782 | No | ||
| 104 | SLC25A27 | 7406 | 1.004 | 0.2800 | No | ||
| 105 | NPPB | 7408 | 1.004 | 0.2818 | No | ||
| 106 | NODAL | 7570 | 0.969 | 0.2763 | No | ||
| 107 | NFATC3 | 7606 | 0.960 | 0.2765 | No | ||
| 108 | GOLGA3 | 7792 | 0.922 | 0.2697 | No | ||
| 109 | PLSCR4 | 7943 | 0.885 | 0.2645 | No | ||
| 110 | NPY2R | 8010 | 0.868 | 0.2631 | No | ||
| 111 | LTC4S | 8034 | 0.862 | 0.2637 | No | ||
| 112 | SLC2A12 | 8132 | 0.839 | 0.2609 | No | ||
| 113 | PIK3R2 | 8374 | 0.789 | 0.2513 | No | ||
| 114 | MEPE | 8743 | 0.702 | 0.2357 | No | ||
| 115 | MYT1 | 8757 | 0.697 | 0.2364 | No | ||
| 116 | FLRT1 | 8823 | 0.681 | 0.2347 | No | ||
| 117 | ACRV1 | 9503 | 0.521 | 0.2045 | No | ||
| 118 | COG7 | 9544 | 0.509 | 0.2036 | No | ||
| 119 | NETO1 | 9631 | 0.487 | 0.2006 | No | ||
| 120 | SCGB1A1 | 9655 | 0.478 | 0.2004 | No | ||
| 121 | TMC5 | 9859 | 0.419 | 0.1919 | No | ||
| 122 | ADAMTS1 | 9890 | 0.412 | 0.1913 | No | ||
| 123 | GLRA2 | 10759 | 0.167 | 0.1517 | No | ||
| 124 | MEGF10 | 10951 | 0.103 | 0.1431 | No | ||
| 125 | GRIN2D | 11214 | 0.025 | 0.1311 | No | ||
| 126 | GJB6 | 11242 | 0.015 | 0.1299 | No | ||
| 127 | TAC4 | 12098 | -0.245 | 0.0910 | No | ||
| 128 | WNT3 | 12450 | -0.349 | 0.0755 | No | ||
| 129 | KLHL10 | 12863 | -0.466 | 0.0574 | No | ||
| 130 | BRD2 | 13031 | -0.511 | 0.0507 | No | ||
| 131 | CDIPT | 13040 | -0.516 | 0.0513 | No | ||
| 132 | IL21R | 13188 | -0.557 | 0.0456 | No | ||
| 133 | PROM1 | 13191 | -0.558 | 0.0466 | No | ||
| 134 | NNAT | 13194 | -0.558 | 0.0476 | No | ||
| 135 | ETV1 | 13308 | -0.590 | 0.0435 | No | ||
| 136 | SPA17 | 13332 | -0.598 | 0.0436 | No | ||
| 137 | MTA2 | 13520 | -0.656 | 0.0362 | No | ||
| 138 | SCN3A | 13546 | -0.661 | 0.0363 | No | ||
| 139 | SLC35C1 | 13712 | -0.703 | 0.0301 | No | ||
| 140 | TLR8 | 13785 | -0.720 | 0.0281 | No | ||
| 141 | MYOD1 | 14057 | -0.794 | 0.0172 | No | ||
| 142 | EMX2 | 14090 | -0.801 | 0.0172 | No | ||
| 143 | C9ORF19 | 14346 | -0.865 | 0.0072 | No | ||
| 144 | TNRC4 | 14348 | -0.866 | 0.0088 | No | ||
| 145 | PPP2R5B | 14506 | -0.909 | 0.0033 | No | ||
| 146 | CAMK2B | 14650 | -0.944 | -0.0015 | No | ||
| 147 | IDH1 | 14882 | -1.010 | -0.0102 | No | ||
| 148 | JAG1 | 14969 | -1.039 | -0.0122 | No | ||
| 149 | CBX3 | 15206 | -1.103 | -0.0209 | No | ||
| 150 | IER5 | 15721 | -1.237 | -0.0422 | No | ||
| 151 | LGI2 | 15735 | -1.240 | -0.0405 | No | ||
| 152 | CAP1 | 15891 | -1.281 | -0.0452 | No | ||
| 153 | GPX1 | 15966 | -1.302 | -0.0461 | No | ||
| 154 | PASK | 15978 | -1.305 | -0.0441 | No | ||
| 155 | EFHD1 | 16250 | -1.375 | -0.0540 | No | ||
| 156 | CNTNAP2 | 16305 | -1.390 | -0.0538 | No | ||
| 157 | SPACA3 | 16328 | -1.398 | -0.0521 | No | ||
| 158 | GABRB3 | 16371 | -1.408 | -0.0514 | No | ||
| 159 | KCNAB1 | 16408 | -1.418 | -0.0503 | No | ||
| 160 | GABRA1 | 16514 | -1.444 | -0.0524 | No | ||
| 161 | PLAT | 16526 | -1.448 | -0.0502 | No | ||
| 162 | BCOR | 16641 | -1.479 | -0.0526 | No | ||
| 163 | APLP2 | 16655 | -1.483 | -0.0504 | No | ||
| 164 | DNAJB5 | 16756 | -1.514 | -0.0521 | No | ||
| 165 | CDK5 | 16995 | -1.585 | -0.0600 | No | ||
| 166 | GAB2 | 17053 | -1.602 | -0.0596 | No | ||
| 167 | LRRC1 | 17126 | -1.623 | -0.0598 | No | ||
| 168 | WBP2 | 17180 | -1.642 | -0.0591 | No | ||
| 169 | GCNT2 | 17244 | -1.662 | -0.0588 | No | ||
| 170 | PON2 | 17330 | -1.693 | -0.0595 | No | ||
| 171 | RIMS2 | 17380 | -1.714 | -0.0585 | No | ||
| 172 | BCL6 | 17488 | -1.746 | -0.0601 | No | ||
| 173 | GRB10 | 17515 | -1.755 | -0.0580 | No | ||
| 174 | UPF2 | 17531 | -1.761 | -0.0553 | No | ||
| 175 | NTF5 | 17983 | -1.915 | -0.0724 | No | ||
| 176 | STAT4 | 18113 | -1.962 | -0.0746 | No | ||
| 177 | ACTA1 | 18569 | -2.146 | -0.0914 | No | ||
| 178 | INVS | 18576 | -2.147 | -0.0876 | No | ||
| 179 | RPS18 | 18620 | -2.168 | -0.0855 | No | ||
| 180 | SEMA6A | 18650 | -2.183 | -0.0827 | No | ||
| 181 | GPR22 | 18818 | -2.256 | -0.0860 | No | ||
| 182 | EWSR1 | 18870 | -2.279 | -0.0840 | No | ||
| 183 | RAB24 | 18950 | -2.315 | -0.0833 | No | ||
| 184 | MAP1LC3B | 19058 | -2.369 | -0.0837 | No | ||
| 185 | ESR1 | 19068 | -2.374 | -0.0796 | No | ||
| 186 | CCDC6 | 19113 | -2.398 | -0.0770 | No | ||
| 187 | NPR1 | 19177 | -2.432 | -0.0753 | No | ||
| 188 | TLK1 | 19196 | -2.443 | -0.0715 | No | ||
| 189 | PCDH8 | 19200 | -2.445 | -0.0669 | No | ||
| 190 | MYOHD1 | 19747 | -2.738 | -0.0868 | No | ||
| 191 | ABCA3 | 19795 | -2.760 | -0.0838 | No | ||
| 192 | RECQL | 20118 | -2.978 | -0.0929 | No | ||
| 193 | DNMT3B | 20249 | -3.058 | -0.0930 | No | ||
| 194 | GTF3C2 | 20261 | -3.065 | -0.0877 | No | ||
| 195 | CYR61 | 20353 | -3.140 | -0.0859 | No | ||
| 196 | UBE2B | 20517 | -3.289 | -0.0872 | No | ||
| 197 | LGI1 | 20542 | -3.300 | -0.0820 | No | ||
| 198 | WASL | 20582 | -3.340 | -0.0774 | No | ||
| 199 | KCNH2 | 20668 | -3.418 | -0.0748 | No | ||
| 200 | CRSP7 | 20747 | -3.506 | -0.0717 | No | ||
| 201 | ATP5I | 20764 | -3.520 | -0.0657 | No | ||
| 202 | SRPK2 | 21253 | -4.263 | -0.0801 | No | ||
| 203 | HSPE1 | 21303 | -4.365 | -0.0740 | No | ||
| 204 | ZDHHC14 | 21422 | -4.658 | -0.0706 | No | ||
| 205 | UTX | 21431 | -4.694 | -0.0620 | No | ||
| 206 | AP3D1 | 21496 | -4.863 | -0.0557 | No | ||
| 207 | KCNQ1 | 21527 | -4.987 | -0.0475 | No | ||
| 208 | PAPOLA | 21551 | -5.071 | -0.0389 | No | ||
| 209 | GNA13 | 21713 | -5.757 | -0.0354 | No | ||
| 210 | SLC4A2 | 21714 | -5.764 | -0.0244 | No | ||
| 211 | ABL1 | 21836 | -7.213 | -0.0162 | No | ||
| 212 | ADRBK1 | 21916 | -11.172 | 0.0014 | No |