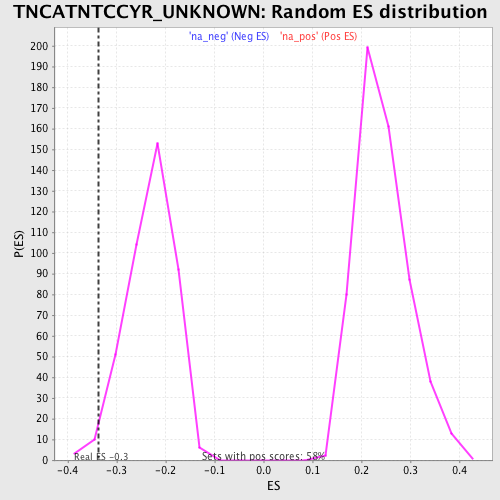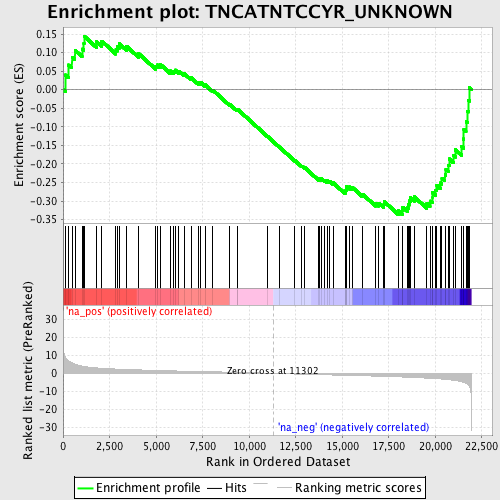
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set01_ATM_minus_versus_SCID |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_neg |
| GeneSet | TNCATNTCCYR_UNKNOWN |
| Enrichment Score (ES) | -0.3362964 |
| Normalized Enrichment Score (NES) | -1.4472411 |
| Nominal p-value | 0.01909308 |
| FDR q-value | 0.3672224 |
| FWER p-Value | 0.98 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | GSPT1 | 122 | 8.818 | 0.0397 | No | ||
| 2 | TTC1 | 300 | 6.828 | 0.0666 | No | ||
| 3 | WHSC1L1 | 477 | 5.742 | 0.0880 | No | ||
| 4 | SNAPC3 | 638 | 4.985 | 0.1062 | No | ||
| 5 | MRPL50 | 1019 | 4.046 | 0.1096 | No | ||
| 6 | SCNM1 | 1118 | 3.864 | 0.1250 | No | ||
| 7 | IPO7 | 1127 | 3.845 | 0.1443 | No | ||
| 8 | FKBPL | 1802 | 3.027 | 0.1290 | No | ||
| 9 | TMEM4 | 2072 | 2.789 | 0.1310 | No | ||
| 10 | COX7C | 2840 | 2.371 | 0.1081 | No | ||
| 11 | WWOX | 2929 | 2.329 | 0.1160 | No | ||
| 12 | FTSJ1 | 3017 | 2.291 | 0.1238 | No | ||
| 13 | CASK | 3409 | 2.138 | 0.1169 | No | ||
| 14 | PSMC3 | 4064 | 1.905 | 0.0968 | No | ||
| 15 | CPEB3 | 4982 | 1.611 | 0.0631 | No | ||
| 16 | CALR3 | 5082 | 1.579 | 0.0666 | No | ||
| 17 | KLF12 | 5243 | 1.532 | 0.0672 | No | ||
| 18 | FOXA3 | 5763 | 1.392 | 0.0506 | No | ||
| 19 | HOXA5 | 5927 | 1.350 | 0.0501 | No | ||
| 20 | DDX25 | 6012 | 1.331 | 0.0531 | No | ||
| 21 | UBE2N | 6220 | 1.280 | 0.0502 | No | ||
| 22 | ERG | 6504 | 1.216 | 0.0434 | No | ||
| 23 | SOX12 | 6877 | 1.128 | 0.0322 | No | ||
| 24 | BDNF | 7264 | 1.037 | 0.0199 | No | ||
| 25 | MRPL54 | 7378 | 1.009 | 0.0199 | No | ||
| 26 | HOXD1 | 7634 | 0.953 | 0.0131 | No | ||
| 27 | MRPL34 | 8052 | 0.858 | -0.0015 | No | ||
| 28 | BET1 | 8952 | 0.649 | -0.0393 | No | ||
| 29 | MTR | 9343 | 0.561 | -0.0543 | No | ||
| 30 | TSTA3 | 9396 | 0.549 | -0.0539 | No | ||
| 31 | APBA3 | 11005 | 0.087 | -0.1269 | No | ||
| 32 | RDH11 | 11621 | -0.103 | -0.1545 | No | ||
| 33 | WNT3 | 12450 | -0.349 | -0.1906 | No | ||
| 34 | STX16 | 12833 | -0.458 | -0.2057 | No | ||
| 35 | GSK3A | 12974 | -0.496 | -0.2096 | No | ||
| 36 | PIK3CG | 13727 | -0.706 | -0.2404 | No | ||
| 37 | CARF | 13765 | -0.715 | -0.2384 | No | ||
| 38 | ADAM22 | 13886 | -0.747 | -0.2400 | No | ||
| 39 | AAMP | 14068 | -0.796 | -0.2442 | No | ||
| 40 | TLE4 | 14190 | -0.826 | -0.2455 | No | ||
| 41 | MLLT7 | 14309 | -0.856 | -0.2465 | No | ||
| 42 | PPP2R5B | 14506 | -0.909 | -0.2509 | No | ||
| 43 | DEF6 | 15154 | -1.088 | -0.2749 | No | ||
| 44 | RASGRP3 | 15191 | -1.098 | -0.2709 | No | ||
| 45 | HDAC8 | 15218 | -1.106 | -0.2664 | No | ||
| 46 | PDE6D | 15234 | -1.111 | -0.2614 | No | ||
| 47 | P2RX3 | 15376 | -1.147 | -0.2619 | No | ||
| 48 | LMO3 | 15539 | -1.191 | -0.2632 | No | ||
| 49 | HSD17B4 | 16098 | -1.333 | -0.2819 | No | ||
| 50 | SMPD3 | 16794 | -1.523 | -0.3059 | No | ||
| 51 | LRRTM3 | 16965 | -1.575 | -0.3056 | No | ||
| 52 | CSMD3 | 17238 | -1.661 | -0.3095 | No | ||
| 53 | SDPR | 17265 | -1.668 | -0.3021 | No | ||
| 54 | CREBL1 | 18013 | -1.924 | -0.3264 | Yes | ||
| 55 | OPHN1 | 18218 | -2.002 | -0.3255 | Yes | ||
| 56 | DDX47 | 18249 | -2.013 | -0.3165 | Yes | ||
| 57 | COVA1 | 18524 | -2.130 | -0.3181 | Yes | ||
| 58 | BIN1 | 18557 | -2.141 | -0.3086 | Yes | ||
| 59 | LMO4 | 18605 | -2.158 | -0.2997 | Yes | ||
| 60 | TLR7 | 18655 | -2.186 | -0.2907 | Yes | ||
| 61 | PRKRIP1 | 18865 | -2.277 | -0.2886 | Yes | ||
| 62 | NT5C1B | 19547 | -2.631 | -0.3063 | Yes | ||
| 63 | SLC35B4 | 19715 | -2.719 | -0.2999 | Yes | ||
| 64 | FEM1A | 19834 | -2.787 | -0.2910 | Yes | ||
| 65 | NTAN1 | 19839 | -2.791 | -0.2769 | Yes | ||
| 66 | DTX2 | 20022 | -2.906 | -0.2703 | Yes | ||
| 67 | EBI2 | 20081 | -2.952 | -0.2578 | Yes | ||
| 68 | PTPN11 | 20285 | -3.088 | -0.2513 | Yes | ||
| 69 | MADD | 20356 | -3.143 | -0.2384 | Yes | ||
| 70 | FYN | 20519 | -3.289 | -0.2289 | Yes | ||
| 71 | MAP2K6 | 20569 | -3.327 | -0.2141 | Yes | ||
| 72 | HMG20A | 20718 | -3.467 | -0.2031 | Yes | ||
| 73 | NR4A3 | 20751 | -3.509 | -0.1865 | Yes | ||
| 74 | DFFA | 20981 | -3.791 | -0.1775 | Yes | ||
| 75 | AGTRAP | 21072 | -3.925 | -0.1615 | Yes | ||
| 76 | SAP130 | 21410 | -4.616 | -0.1533 | Yes | ||
| 77 | CD96 | 21518 | -4.951 | -0.1328 | Yes | ||
| 78 | SYMPK | 21535 | -5.012 | -0.1078 | Yes | ||
| 79 | VPS41 | 21650 | -5.441 | -0.0851 | Yes | ||
| 80 | DPYSL2 | 21727 | -5.920 | -0.0582 | Yes | ||
| 81 | PCQAP | 21771 | -6.214 | -0.0283 | Yes | ||
| 82 | MEN1 | 21827 | -7.073 | 0.0055 | Yes |