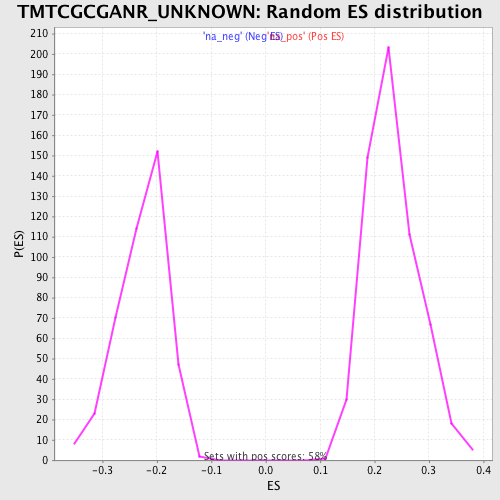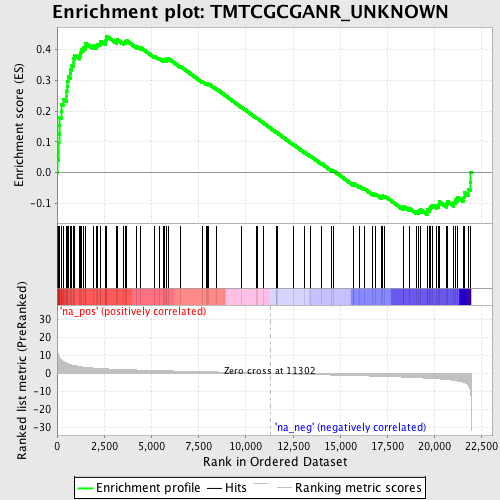
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set01_ATM_minus_versus_SCID |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_pos |
| GeneSet | TMTCGCGANR_UNKNOWN |
| Enrichment Score (ES) | 0.44384667 |
| Normalized Enrichment Score (NES) | 1.910362 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.041897308 |
| FWER p-Value | 0.039 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | SLC25A4 | 17 | 14.011 | 0.0429 | Yes | ||
| 2 | UBE2D3 | 95 | 9.312 | 0.0685 | Yes | ||
| 3 | MATR3 | 98 | 9.240 | 0.0972 | Yes | ||
| 4 | DDX3X | 100 | 9.225 | 0.1260 | Yes | ||
| 5 | DENR | 120 | 8.896 | 0.1529 | Yes | ||
| 6 | HSPA4 | 132 | 8.686 | 0.1795 | Yes | ||
| 7 | EIF5 | 212 | 7.562 | 0.1995 | Yes | ||
| 8 | CDC5L | 227 | 7.452 | 0.2221 | Yes | ||
| 9 | SEC63 | 323 | 6.661 | 0.2385 | Yes | ||
| 10 | ATF2 | 497 | 5.586 | 0.2480 | Yes | ||
| 11 | LUC7L2 | 500 | 5.580 | 0.2654 | Yes | ||
| 12 | HNRPK | 525 | 5.467 | 0.2813 | Yes | ||
| 13 | CHUK | 537 | 5.425 | 0.2977 | Yes | ||
| 14 | RRAGA | 579 | 5.210 | 0.3121 | Yes | ||
| 15 | FGFR1OP2 | 689 | 4.843 | 0.3223 | Yes | ||
| 16 | TLOC1 | 700 | 4.806 | 0.3368 | Yes | ||
| 17 | MAP3K7 | 785 | 4.594 | 0.3473 | Yes | ||
| 18 | TLK2 | 861 | 4.415 | 0.3576 | Yes | ||
| 19 | TTC14 | 865 | 4.407 | 0.3712 | Yes | ||
| 20 | BZW1 | 926 | 4.235 | 0.3817 | Yes | ||
| 21 | PSMD5 | 1197 | 3.724 | 0.3810 | Yes | ||
| 22 | RBPSUH | 1244 | 3.647 | 0.3903 | Yes | ||
| 23 | MBTPS1 | 1274 | 3.605 | 0.4002 | Yes | ||
| 24 | ATP5F1 | 1382 | 3.446 | 0.4060 | Yes | ||
| 25 | POLDIP3 | 1494 | 3.319 | 0.4113 | Yes | ||
| 26 | CCT8 | 1520 | 3.291 | 0.4204 | Yes | ||
| 27 | ZZZ3 | 1908 | 2.935 | 0.4119 | Yes | ||
| 28 | OFD1 | 2086 | 2.777 | 0.4125 | Yes | ||
| 29 | ANAPC4 | 2156 | 2.731 | 0.4178 | Yes | ||
| 30 | ASH2L | 2280 | 2.656 | 0.4205 | Yes | ||
| 31 | MOBKL1A | 2298 | 2.643 | 0.4279 | Yes | ||
| 32 | STXBP4 | 2543 | 2.513 | 0.4246 | Yes | ||
| 33 | NDUFA11 | 2582 | 2.491 | 0.4307 | Yes | ||
| 34 | GAS8 | 2593 | 2.483 | 0.4379 | Yes | ||
| 35 | PFKFB2 | 2633 | 2.462 | 0.4438 | Yes | ||
| 36 | SSR4 | 3165 | 2.234 | 0.4265 | No | ||
| 37 | ZF | 3177 | 2.229 | 0.4330 | No | ||
| 38 | VDAC3 | 3506 | 2.099 | 0.4245 | No | ||
| 39 | RPL17 | 3626 | 2.052 | 0.4255 | No | ||
| 40 | ADNP | 3699 | 2.027 | 0.4285 | No | ||
| 41 | PPP1R12A | 4219 | 1.858 | 0.4105 | No | ||
| 42 | POU4F1 | 4414 | 1.797 | 0.4073 | No | ||
| 43 | TAS1R1 | 5141 | 1.561 | 0.3789 | No | ||
| 44 | PLOD2 | 5407 | 1.488 | 0.3714 | No | ||
| 45 | MDH2 | 5660 | 1.420 | 0.3643 | No | ||
| 46 | FSIP1 | 5707 | 1.406 | 0.3666 | No | ||
| 47 | AGL | 5795 | 1.383 | 0.3669 | No | ||
| 48 | SAP18 | 5817 | 1.378 | 0.3703 | No | ||
| 49 | CD2AP | 5915 | 1.352 | 0.3700 | No | ||
| 50 | ICMT | 6513 | 1.214 | 0.3465 | No | ||
| 51 | RPL12 | 7700 | 0.940 | 0.2952 | No | ||
| 52 | PRDX1 | 7900 | 0.895 | 0.2888 | No | ||
| 53 | RPS19 | 7991 | 0.872 | 0.2874 | No | ||
| 54 | SCAMP5 | 7996 | 0.872 | 0.2900 | No | ||
| 55 | PTPN4 | 8441 | 0.772 | 0.2721 | No | ||
| 56 | THAP1 | 9786 | 0.442 | 0.2119 | No | ||
| 57 | DCTN2 | 10568 | 0.223 | 0.1769 | No | ||
| 58 | KAZALD1 | 10593 | 0.213 | 0.1764 | No | ||
| 59 | UPF3B | 10940 | 0.106 | 0.1609 | No | ||
| 60 | SLC25A11 | 11609 | -0.098 | 0.1307 | No | ||
| 61 | IDH3G | 11620 | -0.103 | 0.1305 | No | ||
| 62 | CCNJ | 11684 | -0.123 | 0.1280 | No | ||
| 63 | LRSAM1 | 12541 | -0.377 | 0.0900 | No | ||
| 64 | NFYC | 13127 | -0.539 | 0.0649 | No | ||
| 65 | BANP | 13442 | -0.632 | 0.0525 | No | ||
| 66 | RPS6 | 13992 | -0.776 | 0.0298 | No | ||
| 67 | TROAP | 14564 | -0.922 | 0.0066 | No | ||
| 68 | RPS7 | 14648 | -0.944 | 0.0057 | No | ||
| 69 | DBR1 | 15687 | -1.230 | -0.0380 | No | ||
| 70 | ANKFY1 | 15698 | -1.232 | -0.0346 | No | ||
| 71 | PIGN | 16004 | -1.312 | -0.0445 | No | ||
| 72 | RPL10A | 16267 | -1.380 | -0.0521 | No | ||
| 73 | COX11 | 16733 | -1.507 | -0.0687 | No | ||
| 74 | RPS15A | 16873 | -1.546 | -0.0703 | No | ||
| 75 | THAP7 | 17208 | -1.652 | -0.0804 | No | ||
| 76 | PURA | 17225 | -1.659 | -0.0759 | No | ||
| 77 | PRDX4 | 17370 | -1.708 | -0.0772 | No | ||
| 78 | BCAS3 | 18347 | -2.057 | -0.1155 | No | ||
| 79 | WDR6 | 18356 | -2.061 | -0.1094 | No | ||
| 80 | RAB33B | 18664 | -2.189 | -0.1166 | No | ||
| 81 | SUV39H1 | 19043 | -2.361 | -0.1265 | No | ||
| 82 | ZFP91 | 19174 | -2.430 | -0.1249 | No | ||
| 83 | SLC12A2 | 19272 | -2.490 | -0.1216 | No | ||
| 84 | ZNF346 | 19604 | -2.662 | -0.1284 | No | ||
| 85 | MAPK14 | 19612 | -2.666 | -0.1204 | No | ||
| 86 | ATF7 | 19732 | -2.732 | -0.1174 | No | ||
| 87 | UHRF2 | 19802 | -2.763 | -0.1119 | No | ||
| 88 | FUS | 19872 | -2.810 | -0.1063 | No | ||
| 89 | ARF3 | 20104 | -2.968 | -0.1076 | No | ||
| 90 | PSMB2 | 20232 | -3.048 | -0.1039 | No | ||
| 91 | LAMP1 | 20255 | -3.062 | -0.0953 | No | ||
| 92 | RPL26 | 20633 | -3.385 | -0.1020 | No | ||
| 93 | NUDT2 | 20688 | -3.438 | -0.0938 | No | ||
| 94 | ASXL1 | 21029 | -3.860 | -0.0973 | No | ||
| 95 | SMARCD2 | 21099 | -3.965 | -0.0881 | No | ||
| 96 | SUPT5H | 21235 | -4.211 | -0.0811 | No | ||
| 97 | CAMK2D | 21559 | -5.093 | -0.0800 | No | ||
| 98 | CNOT3 | 21570 | -5.136 | -0.0644 | No | ||
| 99 | CHURC1 | 21783 | -6.385 | -0.0542 | No | ||
| 100 | DGUOK | 21888 | -8.941 | -0.0311 | No | ||
| 101 | MRP63 | 21913 | -10.802 | 0.0016 | No |