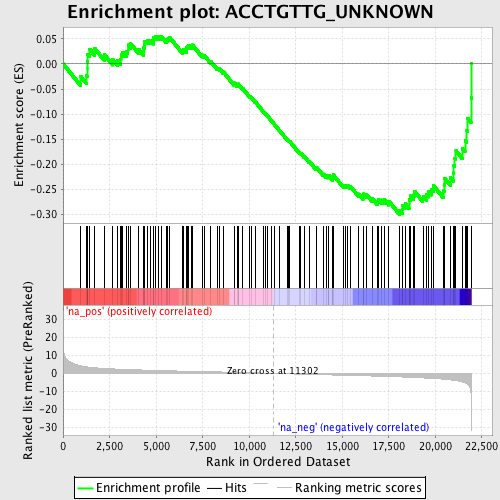
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set01_ATM_minus_versus_SCID |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_neg |
| GeneSet | ACCTGTTG_UNKNOWN |
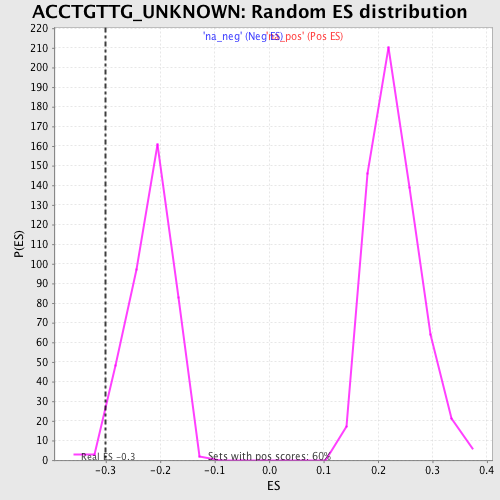
| Enrichment Score (ES) | -0.299649 |
| Normalized Enrichment Score (NES) | -1.3838879 |
| Nominal p-value | 0.01511335 |
| FDR q-value | 0.35548374 |
| FWER p-Value | 0.997 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | ELOVL5 | 932 | 4.218 | -0.0245 | No | ||
| 2 | AP4M1 | 1247 | 3.641 | -0.0231 | No | ||
| 3 | HERC4 | 1294 | 3.564 | -0.0098 | No | ||
| 4 | EPN2 | 1296 | 3.562 | 0.0055 | No | ||
| 5 | PHYHIP | 1328 | 3.517 | 0.0193 | No | ||
| 6 | RSN | 1425 | 3.403 | 0.0296 | No | ||
| 7 | DGCR8 | 1701 | 3.108 | 0.0304 | No | ||
| 8 | RYR1 | 2206 | 2.693 | 0.0189 | No | ||
| 9 | HAP1 | 2669 | 2.447 | 0.0083 | No | ||
| 10 | BCL11A | 2920 | 2.333 | 0.0070 | No | ||
| 11 | AHI1 | 3088 | 2.263 | 0.0091 | No | ||
| 12 | LAMA4 | 3111 | 2.254 | 0.0178 | No | ||
| 13 | GABARAPL2 | 3198 | 2.220 | 0.0235 | No | ||
| 14 | EHD4 | 3390 | 2.144 | 0.0240 | No | ||
| 15 | MRC2 | 3486 | 2.108 | 0.0287 | No | ||
| 16 | SEC24D | 3489 | 2.107 | 0.0377 | No | ||
| 17 | NR3C1 | 3635 | 2.049 | 0.0399 | No | ||
| 18 | NR3C2 | 4057 | 1.907 | 0.0289 | No | ||
| 19 | COL25A1 | 4318 | 1.824 | 0.0249 | No | ||
| 20 | MMP16 | 4325 | 1.821 | 0.0325 | No | ||
| 21 | NKX2-8 | 4381 | 1.804 | 0.0377 | No | ||
| 22 | NPAS2 | 4392 | 1.801 | 0.0450 | No | ||
| 23 | GALR1 | 4511 | 1.766 | 0.0473 | No | ||
| 24 | ZIC4 | 4675 | 1.708 | 0.0472 | No | ||
| 25 | UCHL1 | 4845 | 1.656 | 0.0466 | No | ||
| 26 | FGF17 | 4871 | 1.646 | 0.0525 | No | ||
| 27 | GPR85 | 4972 | 1.616 | 0.0549 | No | ||
| 28 | NOTCH1 | 5134 | 1.563 | 0.0543 | No | ||
| 29 | COX6A2 | 5259 | 1.527 | 0.0552 | No | ||
| 30 | MTUS1 | 5546 | 1.447 | 0.0484 | No | ||
| 31 | KCNQ3 | 5634 | 1.426 | 0.0506 | No | ||
| 32 | SUMO2 | 5716 | 1.403 | 0.0529 | No | ||
| 33 | IGSF4 | 6412 | 1.237 | 0.0264 | No | ||
| 34 | FRAG1 | 6470 | 1.224 | 0.0291 | No | ||
| 35 | JUP | 6615 | 1.192 | 0.0276 | No | ||
| 36 | SLC7A3 | 6621 | 1.191 | 0.0326 | No | ||
| 37 | H2AFY | 6669 | 1.178 | 0.0355 | No | ||
| 38 | FOXB1 | 6745 | 1.160 | 0.0371 | No | ||
| 39 | SESTD1 | 6880 | 1.127 | 0.0358 | No | ||
| 40 | ECEL1 | 6945 | 1.112 | 0.0377 | No | ||
| 41 | KCND2 | 7474 | 0.991 | 0.0178 | No | ||
| 42 | GGN | 7592 | 0.965 | 0.0166 | No | ||
| 43 | KCNV1 | 7937 | 0.887 | 0.0046 | No | ||
| 44 | FAF1 | 8302 | 0.803 | -0.0086 | No | ||
| 45 | NUDT11 | 8397 | 0.784 | -0.0095 | No | ||
| 46 | PCBP4 | 8607 | 0.733 | -0.0159 | No | ||
| 47 | PGM3 | 9182 | 0.598 | -0.0396 | No | ||
| 48 | DOC2A | 9202 | 0.592 | -0.0379 | No | ||
| 49 | SNCA | 9379 | 0.552 | -0.0436 | No | ||
| 50 | YARS | 9400 | 0.548 | -0.0422 | No | ||
| 51 | RWDD2 | 9404 | 0.547 | -0.0399 | No | ||
| 52 | RIMS4 | 9653 | 0.479 | -0.0492 | No | ||
| 53 | GABRE | 10023 | 0.377 | -0.0645 | No | ||
| 54 | OSBPL11 | 10124 | 0.347 | -0.0676 | No | ||
| 55 | ZHX2 | 10314 | 0.296 | -0.0750 | No | ||
| 56 | SHOX2 | 10761 | 0.166 | -0.0947 | No | ||
| 57 | HCRTR1 | 10885 | 0.122 | -0.0998 | No | ||
| 58 | RASGEF1B | 10981 | 0.096 | -0.1037 | No | ||
| 59 | SOX21 | 11175 | 0.037 | -0.1124 | No | ||
| 60 | LRRN3 | 11383 | -0.027 | -0.1218 | No | ||
| 61 | HRH3 | 11624 | -0.104 | -0.1323 | No | ||
| 62 | PNMA5 | 12060 | -0.231 | -0.1512 | No | ||
| 63 | NANOS1 | 12107 | -0.247 | -0.1523 | No | ||
| 64 | MCM7 | 12175 | -0.266 | -0.1542 | No | ||
| 65 | ATP6V1C2 | 12713 | -0.426 | -0.1770 | No | ||
| 66 | HNRPUL1 | 12768 | -0.442 | -0.1775 | No | ||
| 67 | INHBA | 12953 | -0.490 | -0.1838 | No | ||
| 68 | TJP1 | 13243 | -0.571 | -0.1946 | No | ||
| 69 | CASKIN2 | 13589 | -0.672 | -0.2075 | No | ||
| 70 | EML4 | 13614 | -0.675 | -0.2057 | No | ||
| 71 | RERG | 13980 | -0.773 | -0.2191 | No | ||
| 72 | JPH3 | 14131 | -0.812 | -0.2224 | No | ||
| 73 | SORCS3 | 14234 | -0.835 | -0.2235 | No | ||
| 74 | HUNK | 14266 | -0.842 | -0.2213 | No | ||
| 75 | GALNTL2 | 14487 | -0.902 | -0.2275 | No | ||
| 76 | COL18A1 | 14497 | -0.907 | -0.2240 | No | ||
| 77 | RASL10B | 14502 | -0.908 | -0.2202 | No | ||
| 78 | ERF | 15068 | -1.064 | -0.2415 | No | ||
| 79 | MKNK2 | 15165 | -1.091 | -0.2412 | No | ||
| 80 | RAB5C | 15288 | -1.125 | -0.2419 | No | ||
| 81 | MUSK | 15439 | -1.165 | -0.2438 | No | ||
| 82 | RIMS1 | 15889 | -1.281 | -0.2588 | No | ||
| 83 | KCNK13 | 16115 | -1.337 | -0.2633 | No | ||
| 84 | PRRX1 | 16141 | -1.346 | -0.2587 | No | ||
| 85 | CHST11 | 16296 | -1.389 | -0.2597 | No | ||
| 86 | CACNB3 | 16623 | -1.475 | -0.2683 | No | ||
| 87 | FALZ | 16871 | -1.546 | -0.2729 | No | ||
| 88 | GAP43 | 16941 | -1.567 | -0.2693 | No | ||
| 89 | LRRC1 | 17126 | -1.623 | -0.2707 | No | ||
| 90 | CHRNB1 | 17275 | -1.672 | -0.2703 | No | ||
| 91 | KIF13A | 17508 | -1.753 | -0.2734 | No | ||
| 92 | SOX10 | 18083 | -1.952 | -0.2912 | Yes | ||
| 93 | RARB | 18229 | -2.006 | -0.2892 | Yes | ||
| 94 | EYA4 | 18238 | -2.008 | -0.2809 | Yes | ||
| 95 | SCN5A | 18373 | -2.068 | -0.2781 | Yes | ||
| 96 | DDAH2 | 18588 | -2.150 | -0.2786 | Yes | ||
| 97 | PIP5K1B | 18589 | -2.151 | -0.2693 | Yes | ||
| 98 | RAPGEF6 | 18652 | -2.185 | -0.2627 | Yes | ||
| 99 | NUDT10 | 18825 | -2.259 | -0.2608 | Yes | ||
| 100 | NR4A2 | 18880 | -2.284 | -0.2535 | Yes | ||
| 101 | USP30 | 19337 | -2.514 | -0.2635 | Yes | ||
| 102 | ATBF1 | 19505 | -2.609 | -0.2599 | Yes | ||
| 103 | CHCHD3 | 19616 | -2.667 | -0.2534 | Yes | ||
| 104 | ARL4A | 19782 | -2.756 | -0.2490 | Yes | ||
| 105 | ABR | 19899 | -2.828 | -0.2421 | Yes | ||
| 106 | OCRL | 20429 | -3.197 | -0.2526 | Yes | ||
| 107 | PICALM | 20475 | -3.247 | -0.2406 | Yes | ||
| 108 | CSNK1A1 | 20494 | -3.270 | -0.2273 | Yes | ||
| 109 | YPEL1 | 20806 | -3.576 | -0.2261 | Yes | ||
| 110 | MSI2 | 20963 | -3.762 | -0.2170 | Yes | ||
| 111 | KPNA6 | 20997 | -3.815 | -0.2020 | Yes | ||
| 112 | BCL9 | 21051 | -3.896 | -0.1876 | Yes | ||
| 113 | PHF2 | 21108 | -3.985 | -0.1730 | Yes | ||
| 114 | NRF1 | 21445 | -4.729 | -0.1680 | Yes | ||
| 115 | WDR1 | 21600 | -5.239 | -0.1524 | Yes | ||
| 116 | NEURL | 21692 | -5.636 | -0.1322 | Yes | ||
| 117 | NONO | 21732 | -5.944 | -0.1083 | Yes | ||
| 118 | ANXA9 | 21920 | -11.451 | -0.0674 | Yes | ||
| 119 | FOXO3A | 21941 | -15.889 | 0.0003 | Yes |