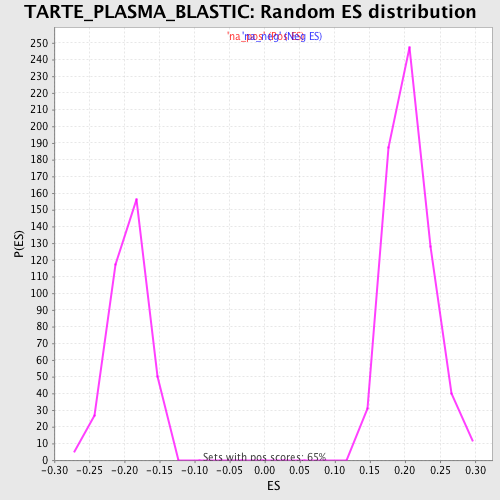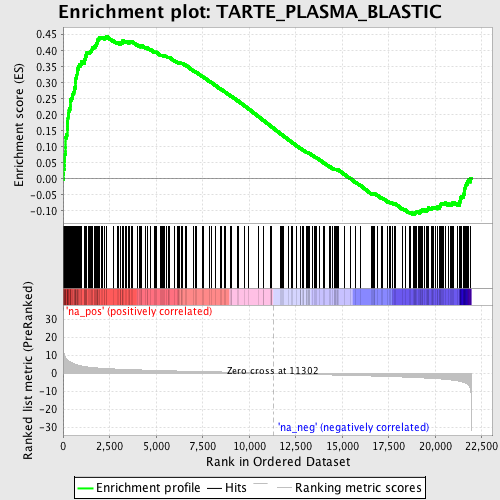
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set01_ATM_minus_versus_SCID |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_pos |
| GeneSet | TARTE_PLASMA_BLASTIC |
| Enrichment Score (ES) | 0.44563895 |
| Normalized Enrichment Score (NES) | 2.1656253 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.0011987356 |
| FWER p-Value | 0.013 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | SFRS10 | 19 | 13.401 | 0.0158 | Yes | ||
| 2 | ABCE1 | 41 | 11.383 | 0.0290 | Yes | ||
| 3 | EZH2 | 58 | 10.502 | 0.0413 | Yes | ||
| 4 | PSMD14 | 67 | 10.195 | 0.0536 | Yes | ||
| 5 | ETF1 | 77 | 9.911 | 0.0655 | Yes | ||
| 6 | UBE2D3 | 95 | 9.312 | 0.0763 | Yes | ||
| 7 | GSPT1 | 122 | 8.818 | 0.0860 | Yes | ||
| 8 | GNL2 | 126 | 8.769 | 0.0968 | Yes | ||
| 9 | GPIAP1 | 131 | 8.703 | 0.1074 | Yes | ||
| 10 | HSPA4 | 132 | 8.686 | 0.1182 | Yes | ||
| 11 | PSME4 | 133 | 8.679 | 0.1290 | Yes | ||
| 12 | NASP | 168 | 8.269 | 0.1377 | Yes | ||
| 13 | IQGAP1 | 211 | 7.562 | 0.1451 | Yes | ||
| 14 | EIF5 | 212 | 7.562 | 0.1545 | Yes | ||
| 15 | DLG7 | 229 | 7.444 | 0.1630 | Yes | ||
| 16 | COPS5 | 237 | 7.333 | 0.1718 | Yes | ||
| 17 | CCT6A | 247 | 7.235 | 0.1804 | Yes | ||
| 18 | CCT2 | 259 | 7.109 | 0.1887 | Yes | ||
| 19 | HPRT1 | 264 | 7.095 | 0.1974 | Yes | ||
| 20 | CYCS | 295 | 6.886 | 0.2045 | Yes | ||
| 21 | VBP1 | 314 | 6.733 | 0.2121 | Yes | ||
| 22 | PRIM1 | 324 | 6.659 | 0.2199 | Yes | ||
| 23 | USP14 | 373 | 6.316 | 0.2256 | Yes | ||
| 24 | RAB27A | 378 | 6.311 | 0.2332 | Yes | ||
| 25 | PPP1R2 | 381 | 6.281 | 0.2409 | Yes | ||
| 26 | CCT5 | 400 | 6.178 | 0.2478 | Yes | ||
| 27 | HUWE1 | 463 | 5.818 | 0.2522 | Yes | ||
| 28 | RARS | 509 | 5.559 | 0.2570 | Yes | ||
| 29 | APPBP1 | 520 | 5.509 | 0.2634 | Yes | ||
| 30 | EIF3S10 | 536 | 5.426 | 0.2694 | Yes | ||
| 31 | KIF2A | 585 | 5.201 | 0.2737 | Yes | ||
| 32 | IGF1 | 591 | 5.167 | 0.2799 | Yes | ||
| 33 | GLIPR1 | 627 | 5.015 | 0.2845 | Yes | ||
| 34 | PAICS | 653 | 4.935 | 0.2895 | Yes | ||
| 35 | TCP1 | 662 | 4.923 | 0.2952 | Yes | ||
| 36 | TCEB1 | 663 | 4.921 | 0.3013 | Yes | ||
| 37 | UBE2L3 | 682 | 4.865 | 0.3065 | Yes | ||
| 38 | CSE1L | 685 | 4.855 | 0.3125 | Yes | ||
| 39 | NARS | 697 | 4.826 | 0.3180 | Yes | ||
| 40 | CLCN3 | 723 | 4.739 | 0.3227 | Yes | ||
| 41 | NKRF | 746 | 4.673 | 0.3275 | Yes | ||
| 42 | CCT4 | 750 | 4.659 | 0.3331 | Yes | ||
| 43 | RRM1 | 763 | 4.636 | 0.3383 | Yes | ||
| 44 | EBNA1BP2 | 779 | 4.608 | 0.3434 | Yes | ||
| 45 | TOPBP1 | 811 | 4.527 | 0.3476 | Yes | ||
| 46 | SMC1A | 837 | 4.454 | 0.3520 | Yes | ||
| 47 | PRPS2 | 860 | 4.418 | 0.3564 | Yes | ||
| 48 | RANBP5 | 953 | 4.185 | 0.3574 | Yes | ||
| 49 | CKAP1 | 985 | 4.116 | 0.3611 | Yes | ||
| 50 | BCL3 | 989 | 4.105 | 0.3660 | Yes | ||
| 51 | IFI30 | 1135 | 3.829 | 0.3641 | Yes | ||
| 52 | MYO5A | 1159 | 3.779 | 0.3677 | Yes | ||
| 53 | ITGB1 | 1163 | 3.777 | 0.3723 | Yes | ||
| 54 | YBX1 | 1188 | 3.732 | 0.3758 | Yes | ||
| 55 | TOP2B | 1204 | 3.715 | 0.3797 | Yes | ||
| 56 | TIMM17A | 1214 | 3.698 | 0.3839 | Yes | ||
| 57 | TFRC | 1245 | 3.642 | 0.3871 | Yes | ||
| 58 | BAG1 | 1255 | 3.631 | 0.3912 | Yes | ||
| 59 | PDAP1 | 1278 | 3.598 | 0.3946 | Yes | ||
| 60 | ATP5F1 | 1382 | 3.446 | 0.3942 | Yes | ||
| 61 | GART | 1443 | 3.379 | 0.3956 | Yes | ||
| 62 | TOP1 | 1486 | 3.326 | 0.3978 | Yes | ||
| 63 | SLC16A1 | 1513 | 3.302 | 0.4007 | Yes | ||
| 64 | NNT | 1551 | 3.261 | 0.4030 | Yes | ||
| 65 | PSMA2 | 1560 | 3.252 | 0.4067 | Yes | ||
| 66 | UBE2S | 1587 | 3.216 | 0.4095 | Yes | ||
| 67 | PLK4 | 1661 | 3.143 | 0.4100 | Yes | ||
| 68 | GGH | 1685 | 3.127 | 0.4129 | Yes | ||
| 69 | LIMS1 | 1719 | 3.096 | 0.4152 | Yes | ||
| 70 | MAPK6 | 1746 | 3.072 | 0.4178 | Yes | ||
| 71 | CDC27 | 1782 | 3.045 | 0.4200 | Yes | ||
| 72 | SOD2 | 1804 | 3.024 | 0.4228 | Yes | ||
| 73 | EIF2S1 | 1825 | 3.000 | 0.4256 | Yes | ||
| 74 | KIF23 | 1835 | 2.992 | 0.4289 | Yes | ||
| 75 | ELMO1 | 1838 | 2.992 | 0.4325 | Yes | ||
| 76 | SNRPB2 | 1870 | 2.965 | 0.4347 | Yes | ||
| 77 | TKT | 1907 | 2.935 | 0.4367 | Yes | ||
| 78 | CCNE1 | 1931 | 2.912 | 0.4393 | Yes | ||
| 79 | UFD1L | 1946 | 2.891 | 0.4422 | Yes | ||
| 80 | UQCRC2 | 2041 | 2.821 | 0.4414 | Yes | ||
| 81 | TARS | 2116 | 2.760 | 0.4414 | Yes | ||
| 82 | GALNT1 | 2227 | 2.683 | 0.4397 | Yes | ||
| 83 | CTPS | 2231 | 2.682 | 0.4429 | Yes | ||
| 84 | MAPK1 | 2313 | 2.637 | 0.4424 | Yes | ||
| 85 | PWP1 | 2315 | 2.637 | 0.4456 | Yes | ||
| 86 | PCNA | 2701 | 2.431 | 0.4309 | No | ||
| 87 | ATIC | 2910 | 2.337 | 0.4242 | No | ||
| 88 | PSMC5 | 2965 | 2.316 | 0.4246 | No | ||
| 89 | PSMA3 | 3100 | 2.259 | 0.4212 | No | ||
| 90 | MRPL19 | 3105 | 2.258 | 0.4238 | No | ||
| 91 | MAD2L1 | 3109 | 2.254 | 0.4265 | No | ||
| 92 | RFC3 | 3176 | 2.229 | 0.4262 | No | ||
| 93 | PSMA1 | 3180 | 2.227 | 0.4288 | No | ||
| 94 | BYSL | 3202 | 2.218 | 0.4306 | No | ||
| 95 | ATP13A3 | 3249 | 2.199 | 0.4312 | No | ||
| 96 | DUT | 3356 | 2.158 | 0.4290 | No | ||
| 97 | ORC1L | 3415 | 2.136 | 0.4290 | No | ||
| 98 | MAPKAPK3 | 3529 | 2.090 | 0.4264 | No | ||
| 99 | ATP5J | 3581 | 2.069 | 0.4266 | No | ||
| 100 | PAK2 | 3589 | 2.066 | 0.4289 | No | ||
| 101 | TMEM97 | 3657 | 2.040 | 0.4283 | No | ||
| 102 | S100A10 | 3710 | 2.023 | 0.4284 | No | ||
| 103 | ARID3A | 3987 | 1.934 | 0.4181 | No | ||
| 104 | PSMD4 | 4092 | 1.897 | 0.4156 | No | ||
| 105 | VDAC1 | 4182 | 1.868 | 0.4139 | No | ||
| 106 | HNRPC | 4226 | 1.855 | 0.4142 | No | ||
| 107 | JARID1A | 4229 | 1.854 | 0.4164 | No | ||
| 108 | ACVR1 | 4432 | 1.789 | 0.4093 | No | ||
| 109 | AHCY | 4519 | 1.764 | 0.4075 | No | ||
| 110 | NDUFV2 | 4522 | 1.764 | 0.4096 | No | ||
| 111 | GARS | 4697 | 1.700 | 0.4037 | No | ||
| 112 | ENTPD1 | 4907 | 1.634 | 0.3961 | No | ||
| 113 | BLMH | 4957 | 1.622 | 0.3958 | No | ||
| 114 | RFC4 | 4995 | 1.606 | 0.3961 | No | ||
| 115 | SNRPD3 | 5251 | 1.529 | 0.3863 | No | ||
| 116 | CCNA2 | 5287 | 1.521 | 0.3865 | No | ||
| 117 | JTV1 | 5351 | 1.503 | 0.3855 | No | ||
| 118 | TMSB10 | 5403 | 1.489 | 0.3850 | No | ||
| 119 | KIF14 | 5441 | 1.477 | 0.3851 | No | ||
| 120 | ATP1B3 | 5534 | 1.451 | 0.3827 | No | ||
| 121 | TRAP1 | 5640 | 1.425 | 0.3796 | No | ||
| 122 | CDKN3 | 5676 | 1.414 | 0.3797 | No | ||
| 123 | SUMO2 | 5716 | 1.403 | 0.3797 | No | ||
| 124 | KLHDC3 | 5983 | 1.336 | 0.3691 | No | ||
| 125 | AIM1 | 6135 | 1.301 | 0.3637 | No | ||
| 126 | CCND2 | 6194 | 1.286 | 0.3626 | No | ||
| 127 | SLC19A1 | 6211 | 1.281 | 0.3635 | No | ||
| 128 | ABCD3 | 6271 | 1.268 | 0.3624 | No | ||
| 129 | PPIF | 6348 | 1.250 | 0.3604 | No | ||
| 130 | TMPO | 6375 | 1.245 | 0.3607 | No | ||
| 131 | MYCBP | 6407 | 1.239 | 0.3609 | No | ||
| 132 | IL2RB | 6551 | 1.206 | 0.3558 | No | ||
| 133 | CDKN1A | 6632 | 1.188 | 0.3535 | No | ||
| 134 | SFRS3 | 7029 | 1.092 | 0.3366 | No | ||
| 135 | CCNB1 | 7091 | 1.075 | 0.3351 | No | ||
| 136 | GLDC | 7191 | 1.053 | 0.3319 | No | ||
| 137 | MCM3 | 7488 | 0.988 | 0.3195 | No | ||
| 138 | MVP | 7551 | 0.976 | 0.3178 | No | ||
| 139 | MRPL12 | 7870 | 0.902 | 0.3043 | No | ||
| 140 | NDUFS1 | 7965 | 0.880 | 0.3010 | No | ||
| 141 | MSN | 8175 | 0.828 | 0.2924 | No | ||
| 142 | HPGD | 8438 | 0.772 | 0.2813 | No | ||
| 143 | NP | 8522 | 0.755 | 0.2784 | No | ||
| 144 | OAT | 8650 | 0.725 | 0.2734 | No | ||
| 145 | CASP1 | 8748 | 0.700 | 0.2698 | No | ||
| 146 | WARS | 8982 | 0.642 | 0.2599 | No | ||
| 147 | POLD2 | 9027 | 0.633 | 0.2586 | No | ||
| 148 | TNFSF7 | 9067 | 0.622 | 0.2576 | No | ||
| 149 | KYNU | 9385 | 0.551 | 0.2436 | No | ||
| 150 | YARS | 9400 | 0.548 | 0.2437 | No | ||
| 151 | MTHFD1 | 9742 | 0.456 | 0.2285 | No | ||
| 152 | PSMC3IP | 9947 | 0.396 | 0.2196 | No | ||
| 153 | CCT7 | 10512 | 0.238 | 0.1939 | No | ||
| 154 | LBR | 10783 | 0.159 | 0.1816 | No | ||
| 155 | BIRC5 | 10791 | 0.157 | 0.1815 | No | ||
| 156 | MPHOSPH6 | 11156 | 0.042 | 0.1647 | No | ||
| 157 | EBP | 11179 | 0.036 | 0.1637 | No | ||
| 158 | RNASE6 | 11210 | 0.028 | 0.1624 | No | ||
| 159 | TUBG1 | 11222 | 0.021 | 0.1619 | No | ||
| 160 | MTHFD2 | 11679 | -0.122 | 0.1410 | No | ||
| 161 | SLC39A7 | 11743 | -0.141 | 0.1383 | No | ||
| 162 | FASN | 11773 | -0.148 | 0.1371 | No | ||
| 163 | TUBA1 | 11825 | -0.163 | 0.1350 | No | ||
| 164 | PRKDC | 12100 | -0.245 | 0.1227 | No | ||
| 165 | EED | 12272 | -0.293 | 0.1151 | No | ||
| 166 | ASNS | 12348 | -0.318 | 0.1121 | No | ||
| 167 | FDPS | 12555 | -0.380 | 0.1030 | No | ||
| 168 | ASNA1 | 12731 | -0.430 | 0.0955 | No | ||
| 169 | ATP5O | 12752 | -0.437 | 0.0951 | No | ||
| 170 | CKS2 | 12769 | -0.442 | 0.0949 | No | ||
| 171 | SSR1 | 12858 | -0.465 | 0.0914 | No | ||
| 172 | GZMB | 12914 | -0.481 | 0.0895 | No | ||
| 173 | ITGB7 | 12915 | -0.481 | 0.0901 | No | ||
| 174 | UQCRH | 13079 | -0.527 | 0.0832 | No | ||
| 175 | SGK | 13093 | -0.530 | 0.0833 | No | ||
| 176 | CD180 | 13125 | -0.538 | 0.0825 | No | ||
| 177 | SNRPF | 13159 | -0.548 | 0.0817 | No | ||
| 178 | RRM2 | 13177 | -0.554 | 0.0816 | No | ||
| 179 | LDHB | 13230 | -0.568 | 0.0799 | No | ||
| 180 | UBE2C | 13259 | -0.576 | 0.0793 | No | ||
| 181 | PDLIM1 | 13401 | -0.622 | 0.0736 | No | ||
| 182 | PSMA5 | 13516 | -0.653 | 0.0691 | No | ||
| 183 | SCAP | 13580 | -0.671 | 0.0671 | No | ||
| 184 | SLC7A5 | 13601 | -0.673 | 0.0670 | No | ||
| 185 | DTYMK | 13771 | -0.716 | 0.0601 | No | ||
| 186 | GALNT3 | 14003 | -0.780 | 0.0504 | No | ||
| 187 | YWHAH | 14065 | -0.795 | 0.0485 | No | ||
| 188 | FLNA | 14301 | -0.852 | 0.0388 | No | ||
| 189 | COX6C | 14341 | -0.864 | 0.0380 | No | ||
| 190 | NEK2 | 14496 | -0.906 | 0.0321 | No | ||
| 191 | TROAP | 14564 | -0.922 | 0.0301 | No | ||
| 192 | SELL | 14591 | -0.928 | 0.0301 | No | ||
| 193 | LMNB1 | 14609 | -0.933 | 0.0304 | No | ||
| 194 | NFIL3 | 14638 | -0.941 | 0.0303 | No | ||
| 195 | NME1 | 14665 | -0.949 | 0.0303 | No | ||
| 196 | FAS | 14741 | -0.969 | 0.0280 | No | ||
| 197 | COASY | 14759 | -0.974 | 0.0285 | No | ||
| 198 | U2AF2 | 14773 | -0.978 | 0.0291 | No | ||
| 199 | EXOSC2 | 15127 | -1.081 | 0.0141 | No | ||
| 200 | TAX1BP3 | 15440 | -1.166 | 0.0012 | No | ||
| 201 | CDC20 | 15732 | -1.240 | -0.0107 | No | ||
| 202 | PSME2 | 15970 | -1.302 | -0.0200 | No | ||
| 203 | KIFC1 | 16592 | -1.467 | -0.0469 | No | ||
| 204 | TUBA3 | 16636 | -1.477 | -0.0470 | No | ||
| 205 | CPOX | 16652 | -1.482 | -0.0459 | No | ||
| 206 | ICT1 | 16706 | -1.499 | -0.0464 | No | ||
| 207 | CKS1B | 16730 | -1.506 | -0.0456 | No | ||
| 208 | ANP32A | 16876 | -1.547 | -0.0504 | No | ||
| 209 | RPA1 | 17082 | -1.611 | -0.0579 | No | ||
| 210 | XRCC6 | 17165 | -1.636 | -0.0596 | No | ||
| 211 | UCHL3 | 17455 | -1.737 | -0.0708 | No | ||
| 212 | LGALS1 | 17533 | -1.763 | -0.0721 | No | ||
| 213 | SDHB | 17604 | -1.785 | -0.0732 | No | ||
| 214 | PSCDBP | 17697 | -1.817 | -0.0751 | No | ||
| 215 | FDFT1 | 17798 | -1.850 | -0.0775 | No | ||
| 216 | CENPE | 17851 | -1.870 | -0.0775 | No | ||
| 217 | IDH2 | 18240 | -2.009 | -0.0929 | No | ||
| 218 | LDLR | 18371 | -2.068 | -0.0964 | No | ||
| 219 | TTK | 18591 | -2.151 | -0.1038 | No | ||
| 220 | ESPL1 | 18665 | -2.189 | -0.1045 | No | ||
| 221 | PFKP | 18804 | -2.250 | -0.1080 | No | ||
| 222 | EWSR1 | 18870 | -2.279 | -0.1082 | No | ||
| 223 | ACTA2 | 18890 | -2.287 | -0.1062 | No | ||
| 224 | DDX39 | 18913 | -2.299 | -0.1044 | No | ||
| 225 | DBI | 18955 | -2.317 | -0.1034 | No | ||
| 226 | MRPL3 | 19010 | -2.344 | -0.1030 | No | ||
| 227 | ARHGDIB | 19075 | -2.378 | -0.1030 | No | ||
| 228 | PTPN1 | 19166 | -2.425 | -0.1041 | No | ||
| 229 | KIF11 | 19203 | -2.448 | -0.1027 | No | ||
| 230 | HMMR | 19211 | -2.453 | -0.1000 | No | ||
| 231 | HADH2 | 19245 | -2.472 | -0.0985 | No | ||
| 232 | FOXK2 | 19288 | -2.495 | -0.0973 | No | ||
| 233 | CD52 | 19301 | -2.500 | -0.0947 | No | ||
| 234 | PSMB8 | 19413 | -2.558 | -0.0967 | No | ||
| 235 | TOP2A | 19518 | -2.617 | -0.0982 | No | ||
| 236 | NDUFB7 | 19559 | -2.638 | -0.0968 | No | ||
| 237 | TRIP13 | 19594 | -2.655 | -0.0951 | No | ||
| 238 | CRADD | 19613 | -2.666 | -0.0926 | No | ||
| 239 | CDC6 | 19624 | -2.669 | -0.0897 | No | ||
| 240 | KIF2C | 19773 | -2.752 | -0.0931 | No | ||
| 241 | GNB1 | 19821 | -2.776 | -0.0919 | No | ||
| 242 | ACOT7 | 19858 | -2.804 | -0.0900 | No | ||
| 243 | AFF1 | 19911 | -2.834 | -0.0889 | No | ||
| 244 | SLA | 19996 | -2.895 | -0.0892 | No | ||
| 245 | DHCR24 | 20099 | -2.965 | -0.0902 | No | ||
| 246 | RECQL | 20118 | -2.978 | -0.0873 | No | ||
| 247 | PSMB2 | 20232 | -3.048 | -0.0888 | No | ||
| 248 | CBX5 | 20242 | -3.054 | -0.0854 | No | ||
| 249 | NDUFA9 | 20264 | -3.066 | -0.0826 | No | ||
| 250 | PRKCB1 | 20298 | -3.099 | -0.0802 | No | ||
| 251 | SUMO1 | 20323 | -3.118 | -0.0775 | No | ||
| 252 | FOXM1 | 20388 | -3.161 | -0.0765 | No | ||
| 253 | TYMS | 20456 | -3.229 | -0.0756 | No | ||
| 254 | UCK2 | 20526 | -3.291 | -0.0747 | No | ||
| 255 | DNAJC7 | 20690 | -3.440 | -0.0779 | No | ||
| 256 | SMARCA4 | 20798 | -3.566 | -0.0784 | No | ||
| 257 | BRD8 | 20892 | -3.675 | -0.0781 | No | ||
| 258 | MRPL23 | 20923 | -3.710 | -0.0749 | No | ||
| 259 | CCNF | 20998 | -3.815 | -0.0736 | No | ||
| 260 | CENPA | 21210 | -4.160 | -0.0782 | No | ||
| 261 | CASP6 | 21299 | -4.360 | -0.0768 | No | ||
| 262 | HSPE1 | 21303 | -4.365 | -0.0715 | No | ||
| 263 | PPP4C | 21328 | -4.405 | -0.0671 | No | ||
| 264 | KIF22 | 21342 | -4.456 | -0.0622 | No | ||
| 265 | RB1 | 21366 | -4.526 | -0.0576 | No | ||
| 266 | ADD1 | 21419 | -4.651 | -0.0543 | No | ||
| 267 | GCLM | 21510 | -4.918 | -0.0523 | No | ||
| 268 | CORO1A | 21531 | -4.999 | -0.0470 | No | ||
| 269 | PAPOLA | 21551 | -5.071 | -0.0416 | No | ||
| 270 | CLIC1 | 21553 | -5.073 | -0.0353 | No | ||
| 271 | IQGAP2 | 21587 | -5.199 | -0.0304 | No | ||
| 272 | PHB | 21598 | -5.236 | -0.0244 | No | ||
| 273 | GIT2 | 21641 | -5.399 | -0.0196 | No | ||
| 274 | OSTF1 | 21652 | -5.451 | -0.0133 | No | ||
| 275 | ZMAT2 | 21718 | -5.796 | -0.0091 | No | ||
| 276 | CDC25B | 21756 | -6.110 | -0.0032 | No | ||
| 277 | ANXA6 | 21898 | -9.615 | 0.0023 | No |