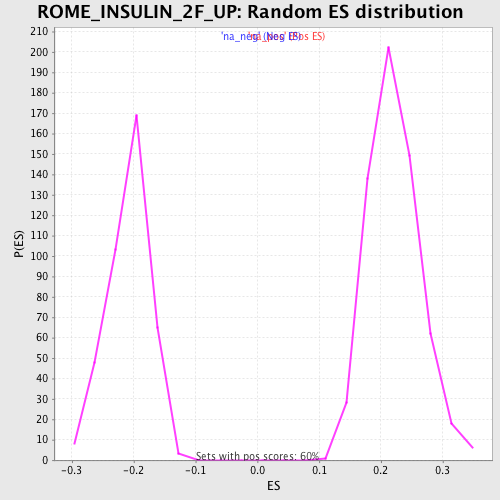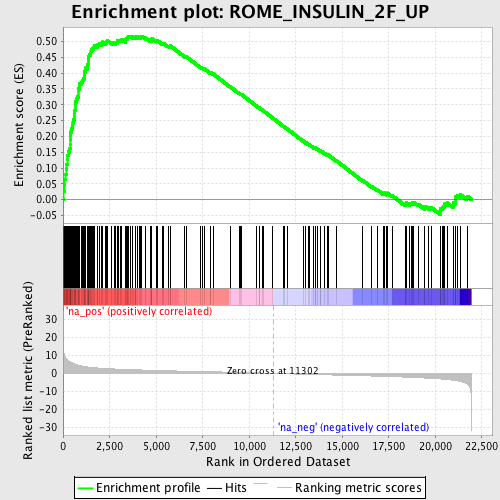
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set01_ATM_minus_versus_SCID |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_pos |
| GeneSet | ROME_INSULIN_2F_UP |
| Enrichment Score (ES) | 0.5169566 |
| Normalized Enrichment Score (NES) | 2.3446085 |
| Nominal p-value | 0.0 |
| FDR q-value | 4.0753168E-4 |
| FWER p-Value | 0.0010 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | RAB5A | 18 | 13.716 | 0.0278 | Yes | ||
| 2 | PSMD14 | 67 | 10.195 | 0.0468 | Yes | ||
| 3 | SET | 97 | 9.283 | 0.0649 | Yes | ||
| 4 | FEM1B | 147 | 8.523 | 0.0804 | Yes | ||
| 5 | SH3GLB1 | 163 | 8.319 | 0.0971 | Yes | ||
| 6 | VCL | 189 | 7.932 | 0.1124 | Yes | ||
| 7 | TPM1 | 232 | 7.421 | 0.1260 | Yes | ||
| 8 | COPS5 | 237 | 7.333 | 0.1411 | Yes | ||
| 9 | ARIH1 | 286 | 6.974 | 0.1534 | Yes | ||
| 10 | ACYP2 | 365 | 6.388 | 0.1632 | Yes | ||
| 11 | AHCYL1 | 376 | 6.313 | 0.1759 | Yes | ||
| 12 | PPP1R2 | 381 | 6.281 | 0.1888 | Yes | ||
| 13 | CANX | 388 | 6.232 | 0.2015 | Yes | ||
| 14 | NPM1 | 398 | 6.184 | 0.2140 | Yes | ||
| 15 | DDX1 | 455 | 5.854 | 0.2237 | Yes | ||
| 16 | SNAPAP | 499 | 5.581 | 0.2333 | Yes | ||
| 17 | HNRPDL | 529 | 5.454 | 0.2434 | Yes | ||
| 18 | EIF2S2 | 565 | 5.270 | 0.2527 | Yes | ||
| 19 | PSMD13 | 603 | 5.104 | 0.2617 | Yes | ||
| 20 | YWHAG | 608 | 5.096 | 0.2721 | Yes | ||
| 21 | CRI1 | 614 | 5.073 | 0.2825 | Yes | ||
| 22 | NRD1 | 657 | 4.931 | 0.2908 | Yes | ||
| 23 | GLRX | 661 | 4.924 | 0.3010 | Yes | ||
| 24 | MTCH2 | 680 | 4.869 | 0.3103 | Yes | ||
| 25 | CLCN3 | 723 | 4.739 | 0.3183 | Yes | ||
| 26 | PGRMC2 | 771 | 4.620 | 0.3257 | Yes | ||
| 27 | GOT2 | 827 | 4.480 | 0.3326 | Yes | ||
| 28 | GSTA4 | 829 | 4.474 | 0.3418 | Yes | ||
| 29 | WBSCR1 | 832 | 4.467 | 0.3511 | Yes | ||
| 30 | UBE2V2 | 877 | 4.381 | 0.3582 | Yes | ||
| 31 | UBQLN1 | 890 | 4.340 | 0.3667 | Yes | ||
| 32 | YWHAQ | 965 | 4.155 | 0.3719 | Yes | ||
| 33 | PTP4A1 | 1028 | 4.027 | 0.3775 | Yes | ||
| 34 | TAF7 | 1074 | 3.936 | 0.3836 | Yes | ||
| 35 | IPO7 | 1127 | 3.845 | 0.3893 | Yes | ||
| 36 | OXA1L | 1138 | 3.822 | 0.3968 | Yes | ||
| 37 | ARCN1 | 1141 | 3.819 | 0.4047 | Yes | ||
| 38 | ST13 | 1199 | 3.718 | 0.4098 | Yes | ||
| 39 | PSMB4 | 1225 | 3.687 | 0.4163 | Yes | ||
| 40 | ARF4 | 1319 | 3.529 | 0.4194 | Yes | ||
| 41 | RNF10 | 1326 | 3.522 | 0.4265 | Yes | ||
| 42 | UBE2I | 1345 | 3.501 | 0.4330 | Yes | ||
| 43 | MORF4L2 | 1347 | 3.499 | 0.4402 | Yes | ||
| 44 | CLNS1A | 1355 | 3.479 | 0.4472 | Yes | ||
| 45 | PSMA4 | 1379 | 3.447 | 0.4533 | Yes | ||
| 46 | USP16 | 1393 | 3.436 | 0.4599 | Yes | ||
| 47 | PTMA | 1471 | 3.346 | 0.4633 | Yes | ||
| 48 | TIMP3 | 1497 | 3.318 | 0.4691 | Yes | ||
| 49 | CCT8 | 1520 | 3.291 | 0.4749 | Yes | ||
| 50 | CFL2 | 1577 | 3.229 | 0.4791 | Yes | ||
| 51 | LARGE | 1653 | 3.150 | 0.4822 | Yes | ||
| 52 | NCKAP1 | 1676 | 3.134 | 0.4877 | Yes | ||
| 53 | PSMA6 | 1834 | 2.997 | 0.4868 | Yes | ||
| 54 | SNRPB2 | 1870 | 2.965 | 0.4914 | Yes | ||
| 55 | PRDX6 | 1942 | 2.895 | 0.4941 | Yes | ||
| 56 | OSR1 | 2071 | 2.789 | 0.4941 | Yes | ||
| 57 | USP9X | 2101 | 2.767 | 0.4985 | Yes | ||
| 58 | UBE3A | 2266 | 2.664 | 0.4966 | Yes | ||
| 59 | SLC35B1 | 2343 | 2.621 | 0.4985 | Yes | ||
| 60 | PAIP2 | 2387 | 2.594 | 0.5020 | Yes | ||
| 61 | BTF3 | 2607 | 2.478 | 0.4971 | Yes | ||
| 62 | SYNPO2L | 2766 | 2.404 | 0.4948 | Yes | ||
| 63 | ARPP-19 | 2833 | 2.374 | 0.4968 | Yes | ||
| 64 | AKAP6 | 2900 | 2.341 | 0.4986 | Yes | ||
| 65 | CAPN2 | 2917 | 2.334 | 0.5028 | Yes | ||
| 66 | SRP14 | 2988 | 2.306 | 0.5043 | Yes | ||
| 67 | PSMA3 | 3100 | 2.259 | 0.5040 | Yes | ||
| 68 | MAT2A | 3156 | 2.238 | 0.5061 | Yes | ||
| 69 | SCOC | 3365 | 2.154 | 0.5010 | Yes | ||
| 70 | SLC39A14 | 3367 | 2.153 | 0.5055 | Yes | ||
| 71 | HDGF | 3378 | 2.149 | 0.5095 | Yes | ||
| 72 | SERPINF1 | 3464 | 2.120 | 0.5100 | Yes | ||
| 73 | ATP6V1E1 | 3481 | 2.111 | 0.5137 | Yes | ||
| 74 | BCL2L2 | 3519 | 2.096 | 0.5164 | Yes | ||
| 75 | VIM | 3632 | 2.050 | 0.5155 | Yes | ||
| 76 | UGP2 | 3747 | 2.011 | 0.5145 | Yes | ||
| 77 | ALDOA | 3888 | 1.961 | 0.5121 | Yes | ||
| 78 | PXMP3 | 3913 | 1.954 | 0.5151 | Yes | ||
| 79 | UACA | 4014 | 1.921 | 0.5145 | Yes | ||
| 80 | ASCL1 | 4117 | 1.889 | 0.5138 | Yes | ||
| 81 | VDAC1 | 4182 | 1.868 | 0.5147 | Yes | ||
| 82 | PPP1R12A | 4219 | 1.858 | 0.5170 | Yes | ||
| 83 | EEF1G | 4425 | 1.792 | 0.5113 | No | ||
| 84 | CDK2AP1 | 4669 | 1.709 | 0.5037 | No | ||
| 85 | STARD7 | 4742 | 1.686 | 0.5039 | No | ||
| 86 | IL13RA1 | 4757 | 1.681 | 0.5068 | No | ||
| 87 | CD36 | 4775 | 1.675 | 0.5095 | No | ||
| 88 | UBE2E3 | 5020 | 1.599 | 0.5016 | No | ||
| 89 | LAPTM4A | 5059 | 1.587 | 0.5032 | No | ||
| 90 | JTV1 | 5351 | 1.503 | 0.4929 | No | ||
| 91 | ITGAE | 5396 | 1.490 | 0.4940 | No | ||
| 92 | GMPR | 5651 | 1.423 | 0.4853 | No | ||
| 93 | CSDA | 5747 | 1.397 | 0.4839 | No | ||
| 94 | ADHFE1 | 5762 | 1.392 | 0.4862 | No | ||
| 95 | BUB1 | 6545 | 1.207 | 0.4528 | No | ||
| 96 | ADFP | 6603 | 1.196 | 0.4527 | No | ||
| 97 | CAMLG | 7398 | 1.006 | 0.4183 | No | ||
| 98 | HSF2 | 7512 | 0.983 | 0.4152 | No | ||
| 99 | GPSN2 | 7579 | 0.967 | 0.4141 | No | ||
| 100 | PRDX1 | 7900 | 0.895 | 0.4013 | No | ||
| 101 | TERF2IP | 7919 | 0.890 | 0.4024 | No | ||
| 102 | AHSA1 | 8059 | 0.856 | 0.3978 | No | ||
| 103 | PAM | 8989 | 0.641 | 0.3564 | No | ||
| 104 | HK2 | 9456 | 0.535 | 0.3362 | No | ||
| 105 | CMAS | 9545 | 0.509 | 0.3332 | No | ||
| 106 | SDCBP | 9591 | 0.495 | 0.3322 | No | ||
| 107 | IHPK3 | 10392 | 0.274 | 0.2960 | No | ||
| 108 | LMCD1 | 10541 | 0.231 | 0.2897 | No | ||
| 109 | RPL7A | 10550 | 0.229 | 0.2898 | No | ||
| 110 | GTF2A2 | 10711 | 0.183 | 0.2828 | No | ||
| 111 | NACA | 10740 | 0.176 | 0.2819 | No | ||
| 112 | C1S | 11266 | 0.009 | 0.2578 | No | ||
| 113 | IFIT1 | 11855 | -0.174 | 0.2312 | No | ||
| 114 | ADSL | 11915 | -0.188 | 0.2289 | No | ||
| 115 | TSG101 | 12081 | -0.239 | 0.2218 | No | ||
| 116 | MAOB | 12893 | -0.475 | 0.1856 | No | ||
| 117 | NT5C3 | 13048 | -0.518 | 0.1796 | No | ||
| 118 | USP22 | 13178 | -0.554 | 0.1748 | No | ||
| 119 | LDHB | 13230 | -0.568 | 0.1737 | No | ||
| 120 | FNTA | 13475 | -0.640 | 0.1638 | No | ||
| 121 | TRIM32 | 13538 | -0.660 | 0.1623 | No | ||
| 122 | DDX48 | 13541 | -0.661 | 0.1636 | No | ||
| 123 | RPLP1 | 13645 | -0.682 | 0.1603 | No | ||
| 124 | EPM2A | 13809 | -0.725 | 0.1543 | No | ||
| 125 | LSM3 | 13827 | -0.731 | 0.1551 | No | ||
| 126 | YWHAH | 14065 | -0.795 | 0.1459 | No | ||
| 127 | RPL41 | 14180 | -0.824 | 0.1423 | No | ||
| 128 | LAPTM4B | 14237 | -0.836 | 0.1415 | No | ||
| 129 | RPS20 | 14667 | -0.949 | 0.1238 | No | ||
| 130 | DAG1 | 16109 | -1.335 | 0.0604 | No | ||
| 131 | PDE4D | 16569 | -1.458 | 0.0424 | No | ||
| 132 | NFE2L1 | 16897 | -1.552 | 0.0306 | No | ||
| 133 | FHL3 | 17200 | -1.648 | 0.0202 | No | ||
| 134 | NDUFS4 | 17253 | -1.666 | 0.0213 | No | ||
| 135 | DPYSL3 | 17398 | -1.718 | 0.0183 | No | ||
| 136 | PLN | 17415 | -1.725 | 0.0211 | No | ||
| 137 | GNPAT | 17678 | -1.810 | 0.0129 | No | ||
| 138 | HERPUD1 | 18392 | -2.075 | -0.0155 | No | ||
| 139 | SLC25A12 | 18422 | -2.087 | -0.0125 | No | ||
| 140 | MRPS16 | 18438 | -2.093 | -0.0088 | No | ||
| 141 | ALS2CR2 | 18634 | -2.178 | -0.0133 | No | ||
| 142 | PUM2 | 18692 | -2.203 | -0.0113 | No | ||
| 143 | ATP6V0B | 18768 | -2.230 | -0.0101 | No | ||
| 144 | RBX1 | 18816 | -2.255 | -0.0075 | No | ||
| 145 | FBP2 | 19085 | -2.382 | -0.0149 | No | ||
| 146 | VPS29 | 19425 | -2.561 | -0.0251 | No | ||
| 147 | PURB | 19434 | -2.565 | -0.0201 | No | ||
| 148 | TALDO1 | 19646 | -2.679 | -0.0242 | No | ||
| 149 | GBAS | 19769 | -2.751 | -0.0241 | No | ||
| 150 | ASH1L | 20267 | -3.070 | -0.0405 | No | ||
| 151 | SORBS1 | 20271 | -3.073 | -0.0342 | No | ||
| 152 | PPP2R3A | 20290 | -3.092 | -0.0286 | No | ||
| 153 | ACTB | 20378 | -3.156 | -0.0260 | No | ||
| 154 | SPARCL1 | 20442 | -3.214 | -0.0222 | No | ||
| 155 | ANXA7 | 20472 | -3.244 | -0.0168 | No | ||
| 156 | UBE2B | 20517 | -3.289 | -0.0119 | No | ||
| 157 | GCN1L1 | 20638 | -3.389 | -0.0104 | No | ||
| 158 | NFIA | 20953 | -3.751 | -0.0169 | No | ||
| 159 | MSI2 | 20963 | -3.762 | -0.0095 | No | ||
| 160 | SPOP | 21064 | -3.916 | -0.0059 | No | ||
| 161 | QARS | 21100 | -3.966 | 0.0007 | No | ||
| 162 | H2AFZ | 21106 | -3.980 | 0.0088 | No | ||
| 163 | STS | 21180 | -4.101 | 0.0140 | No | ||
| 164 | HECTD1 | 21343 | -4.458 | 0.0159 | No | ||
| 165 | COX7A2 | 21701 | -5.668 | 0.0113 | No |