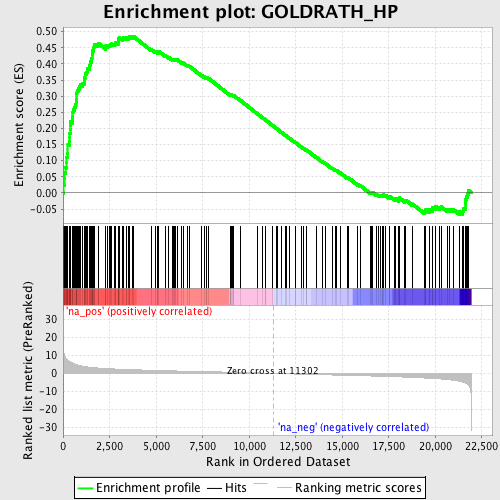
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set01_ATM_minus_versus_SCID |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_pos |
| GeneSet | GOLDRATH_HP |
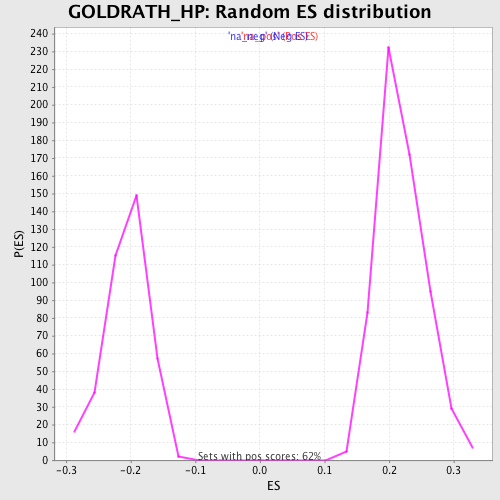
| Enrichment Score (ES) | 0.48622906 |
| Normalized Enrichment Score (NES) | 2.2249699 |
| Nominal p-value | 0.0 |
| FDR q-value | 3.4931288E-4 |
| FWER p-Value | 0.0020 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | CACYBP | 20 | 13.387 | 0.0260 | Yes | ||
| 2 | EIF3S1 | 57 | 10.656 | 0.0458 | Yes | ||
| 3 | SET | 97 | 9.283 | 0.0626 | Yes | ||
| 4 | GNPNAT1 | 134 | 8.666 | 0.0784 | Yes | ||
| 5 | PSMC6 | 160 | 8.356 | 0.0940 | Yes | ||
| 6 | PTGES3 | 164 | 8.319 | 0.1106 | Yes | ||
| 7 | 2610005L07RIK | 214 | 7.556 | 0.1236 | Yes | ||
| 8 | RAD23B | 258 | 7.120 | 0.1359 | Yes | ||
| 9 | CCT2 | 259 | 7.109 | 0.1502 | Yes | ||
| 10 | GMFB | 325 | 6.645 | 0.1606 | Yes | ||
| 11 | ZHX1 | 353 | 6.444 | 0.1723 | Yes | ||
| 12 | JOSD3 | 359 | 6.404 | 0.1849 | Yes | ||
| 13 | DHX9 | 379 | 6.306 | 0.1967 | Yes | ||
| 14 | CANX | 388 | 6.232 | 0.2089 | Yes | ||
| 15 | CETN3 | 402 | 6.168 | 0.2207 | Yes | ||
| 16 | 2700007P21RIK | 505 | 5.567 | 0.2272 | Yes | ||
| 17 | LIN7C | 514 | 5.538 | 0.2380 | Yes | ||
| 18 | SEL1H | 523 | 5.498 | 0.2487 | Yes | ||
| 19 | TM9SF3 | 534 | 5.431 | 0.2591 | Yes | ||
| 20 | CRI1 | 614 | 5.073 | 0.2657 | Yes | ||
| 21 | HRB | 644 | 4.951 | 0.2743 | Yes | ||
| 22 | PB1 | 693 | 4.835 | 0.2818 | Yes | ||
| 23 | TLOC1 | 700 | 4.806 | 0.2912 | Yes | ||
| 24 | SMN1 | 711 | 4.780 | 0.3004 | Yes | ||
| 25 | RAB18 | 713 | 4.768 | 0.3099 | Yes | ||
| 26 | C1QB | 747 | 4.672 | 0.3178 | Yes | ||
| 27 | APP | 805 | 4.540 | 0.3243 | Yes | ||
| 28 | PRPS2 | 860 | 4.418 | 0.3307 | Yes | ||
| 29 | KCTD12 | 912 | 4.267 | 0.3369 | Yes | ||
| 30 | TGOLN1 | 1056 | 3.970 | 0.3383 | Yes | ||
| 31 | IPO7 | 1127 | 3.845 | 0.3428 | Yes | ||
| 32 | UBQLN2 | 1133 | 3.831 | 0.3503 | Yes | ||
| 33 | TAX1BP1 | 1161 | 3.778 | 0.3567 | Yes | ||
| 34 | DYNLL2 | 1179 | 3.744 | 0.3634 | Yes | ||
| 35 | CTNND2 | 1180 | 3.742 | 0.3709 | Yes | ||
| 36 | PDAP1 | 1278 | 3.598 | 0.3737 | Yes | ||
| 37 | TRNT1 | 1312 | 3.537 | 0.3793 | Yes | ||
| 38 | YIPF5 | 1320 | 3.529 | 0.3861 | Yes | ||
| 39 | MKI67IP | 1408 | 3.422 | 0.3890 | Yes | ||
| 40 | TAF9 | 1412 | 3.417 | 0.3957 | Yes | ||
| 41 | QDPR | 1455 | 3.362 | 0.4005 | Yes | ||
| 42 | NUDCD2 | 1456 | 3.359 | 0.4073 | Yes | ||
| 43 | AI788669 | 1514 | 3.297 | 0.4113 | Yes | ||
| 44 | YME1L1 | 1519 | 3.292 | 0.4177 | Yes | ||
| 45 | PRKCA | 1554 | 3.257 | 0.4227 | Yes | ||
| 46 | ITGB1BP1 | 1555 | 3.257 | 0.4292 | Yes | ||
| 47 | BZW2 | 1589 | 3.215 | 0.4342 | Yes | ||
| 48 | BTBD14A | 1597 | 3.206 | 0.4403 | Yes | ||
| 49 | AU021838 | 1640 | 3.163 | 0.4447 | Yes | ||
| 50 | SFPQ | 1655 | 3.149 | 0.4504 | Yes | ||
| 51 | AKAP8 | 1664 | 3.142 | 0.4564 | Yes | ||
| 52 | CPSF2 | 1696 | 3.111 | 0.4612 | Yes | ||
| 53 | RNF11 | 1877 | 2.961 | 0.4589 | Yes | ||
| 54 | ROCK1 | 1902 | 2.938 | 0.4637 | Yes | ||
| 55 | PTDSS1 | 2301 | 2.640 | 0.4507 | Yes | ||
| 56 | RNF141 | 2302 | 2.639 | 0.4560 | Yes | ||
| 57 | GLUL | 2402 | 2.587 | 0.4567 | Yes | ||
| 58 | PDCD2 | 2477 | 2.553 | 0.4584 | Yes | ||
| 59 | ZRSR2 | 2544 | 2.513 | 0.4604 | Yes | ||
| 60 | GOLPH4 | 2619 | 2.469 | 0.4620 | Yes | ||
| 61 | VDAC2 | 2740 | 2.418 | 0.4614 | Yes | ||
| 62 | PCCB | 2799 | 2.389 | 0.4635 | Yes | ||
| 63 | SNX9 | 2824 | 2.375 | 0.4672 | Yes | ||
| 64 | VAMP4 | 2953 | 2.320 | 0.4659 | Yes | ||
| 65 | PSMC5 | 2965 | 2.316 | 0.4701 | Yes | ||
| 66 | TPD52 | 2977 | 2.313 | 0.4742 | Yes | ||
| 67 | TIAL1 | 2987 | 2.307 | 0.4785 | Yes | ||
| 68 | GYK | 3011 | 2.293 | 0.4820 | Yes | ||
| 69 | BYSL | 3202 | 2.218 | 0.4778 | Yes | ||
| 70 | PPA1 | 3224 | 2.213 | 0.4812 | Yes | ||
| 71 | HDGF | 3378 | 2.149 | 0.4785 | Yes | ||
| 72 | MUT | 3387 | 2.146 | 0.4825 | Yes | ||
| 73 | SORL1 | 3522 | 2.093 | 0.4805 | Yes | ||
| 74 | CREB1 | 3579 | 2.070 | 0.4821 | Yes | ||
| 75 | ATP5J | 3581 | 2.069 | 0.4862 | Yes | ||
| 76 | MSL2L1 | 3748 | 2.010 | 0.4826 | No | ||
| 77 | KLF9 | 3801 | 1.991 | 0.4843 | No | ||
| 78 | ATP11A | 4738 | 1.686 | 0.4447 | No | ||
| 79 | 1110008F13RIK | 4943 | 1.623 | 0.4385 | No | ||
| 80 | PLEKHA1 | 5092 | 1.575 | 0.4349 | No | ||
| 81 | SFXN1 | 5096 | 1.573 | 0.4379 | No | ||
| 82 | CSPG3 | 5144 | 1.560 | 0.4389 | No | ||
| 83 | D1ERTD622E | 5482 | 1.465 | 0.4264 | No | ||
| 84 | E2F1 | 5666 | 1.419 | 0.4208 | No | ||
| 85 | PDCD6 | 5900 | 1.357 | 0.4129 | No | ||
| 86 | NOLA2 | 5953 | 1.344 | 0.4132 | No | ||
| 87 | MAGOH | 5979 | 1.338 | 0.4147 | No | ||
| 88 | MYO6 | 6056 | 1.322 | 0.4139 | No | ||
| 89 | AW146242 | 6126 | 1.303 | 0.4133 | No | ||
| 90 | STRN | 6374 | 1.246 | 0.4045 | No | ||
| 91 | EBF3 | 6461 | 1.227 | 0.4030 | No | ||
| 92 | H2AFY | 6669 | 1.178 | 0.3959 | No | ||
| 93 | 2310016A09RIK | 6779 | 1.152 | 0.3932 | No | ||
| 94 | D13WSU177E | 7448 | 0.996 | 0.3645 | No | ||
| 95 | NUDT19 | 7585 | 0.966 | 0.3602 | No | ||
| 96 | SEMA4F | 7686 | 0.942 | 0.3575 | No | ||
| 97 | AQP9 | 7703 | 0.939 | 0.3586 | No | ||
| 98 | IGBP1 | 7827 | 0.912 | 0.3548 | No | ||
| 99 | RPIA | 8988 | 0.641 | 0.3028 | No | ||
| 100 | ENPEP | 9003 | 0.638 | 0.3035 | No | ||
| 101 | EOMES | 9008 | 0.637 | 0.3046 | No | ||
| 102 | SOS2 | 9029 | 0.633 | 0.3049 | No | ||
| 103 | VTI1B | 9109 | 0.613 | 0.3025 | No | ||
| 104 | ITFG1 | 9161 | 0.603 | 0.3014 | No | ||
| 105 | 4833442J19RIK | 9167 | 0.601 | 0.3024 | No | ||
| 106 | CMAS | 9545 | 0.509 | 0.2861 | No | ||
| 107 | UNG | 10452 | 0.255 | 0.2450 | No | ||
| 108 | 2310056P07RIK | 10718 | 0.182 | 0.2332 | No | ||
| 109 | 4933428G09RIK | 10878 | 0.124 | 0.2261 | No | ||
| 110 | ADAM3A | 10894 | 0.120 | 0.2257 | No | ||
| 111 | PNPLA1 | 11253 | 0.012 | 0.2092 | No | ||
| 112 | ZFP131 | 11457 | -0.052 | 0.2000 | No | ||
| 113 | BAG2 | 11526 | -0.074 | 0.1971 | No | ||
| 114 | BRP44 | 11726 | -0.135 | 0.1882 | No | ||
| 115 | IFT88 | 11944 | -0.196 | 0.1786 | No | ||
| 116 | X83328 | 11988 | -0.207 | 0.1770 | No | ||
| 117 | ALDH3A2 | 12172 | -0.266 | 0.1692 | No | ||
| 118 | RECK | 12463 | -0.352 | 0.1566 | No | ||
| 119 | SNRPE | 12480 | -0.358 | 0.1565 | No | ||
| 120 | CYB5R1 | 12824 | -0.456 | 0.1417 | No | ||
| 121 | SDHC | 12906 | -0.478 | 0.1390 | No | ||
| 122 | SOCS3 | 13073 | -0.525 | 0.1324 | No | ||
| 123 | 2310010J17RIK | 13077 | -0.526 | 0.1333 | No | ||
| 124 | SLC7A5 | 13601 | -0.673 | 0.1106 | No | ||
| 125 | PELO | 13932 | -0.760 | 0.0970 | No | ||
| 126 | STT3A | 14077 | -0.798 | 0.0920 | No | ||
| 127 | IRAK1 | 14459 | -0.896 | 0.0763 | No | ||
| 128 | IPP | 14626 | -0.937 | 0.0706 | No | ||
| 129 | NME1 | 14665 | -0.949 | 0.0707 | No | ||
| 130 | AU016916 | 14877 | -1.009 | 0.0630 | No | ||
| 131 | RAB5C | 15288 | -1.125 | 0.0465 | No | ||
| 132 | HSD3B5 | 15339 | -1.138 | 0.0465 | No | ||
| 133 | C330012H03RIK | 15823 | -1.266 | 0.0268 | No | ||
| 134 | GPX1 | 15966 | -1.302 | 0.0229 | No | ||
| 135 | KCNS2 | 16494 | -1.440 | 0.0016 | No | ||
| 136 | F2RL1 | 16590 | -1.467 | 0.0002 | No | ||
| 137 | PARK7 | 16633 | -1.477 | 0.0012 | No | ||
| 138 | MRPS33 | 16856 | -1.541 | -0.0059 | No | ||
| 139 | IMPA2 | 16940 | -1.566 | -0.0065 | No | ||
| 140 | CD55 | 17034 | -1.595 | -0.0076 | No | ||
| 141 | 1110049G11RIK | 17134 | -1.625 | -0.0089 | No | ||
| 142 | IL10RB | 17183 | -1.643 | -0.0078 | No | ||
| 143 | SRPR | 17204 | -1.650 | -0.0054 | No | ||
| 144 | PCGF2 | 17318 | -1.689 | -0.0072 | No | ||
| 145 | E330034G19RIK | 17553 | -1.769 | -0.0144 | No | ||
| 146 | 5730408K05RIK | 17561 | -1.773 | -0.0111 | No | ||
| 147 | NDUFB3 | 17803 | -1.851 | -0.0185 | No | ||
| 148 | CHEK2 | 17844 | -1.867 | -0.0166 | No | ||
| 149 | NDUFC1 | 18039 | -1.935 | -0.0216 | No | ||
| 150 | GALK1 | 18053 | -1.942 | -0.0183 | No | ||
| 151 | ATP5D | 18065 | -1.946 | -0.0149 | No | ||
| 152 | 4930453N24RIK | 18363 | -2.064 | -0.0244 | No | ||
| 153 | 2700094K13RIK | 18416 | -2.085 | -0.0226 | No | ||
| 154 | ATP6V0B | 18768 | -2.230 | -0.0342 | No | ||
| 155 | SCD5 | 19401 | -2.552 | -0.0581 | No | ||
| 156 | MRPS25 | 19466 | -2.587 | -0.0558 | No | ||
| 157 | NFKBIZ | 19493 | -2.604 | -0.0518 | No | ||
| 158 | NDUFAB1 | 19688 | -2.703 | -0.0553 | No | ||
| 159 | CDCA7L | 19695 | -2.706 | -0.0501 | No | ||
| 160 | NTAN1 | 19839 | -2.791 | -0.0511 | No | ||
| 161 | GTPBP1 | 19856 | -2.803 | -0.0462 | No | ||
| 162 | FUBP1 | 19987 | -2.882 | -0.0463 | No | ||
| 163 | NDUFB2 | 20003 | -2.899 | -0.0412 | No | ||
| 164 | 1110059E24RIK | 20248 | -3.057 | -0.0463 | No | ||
| 165 | UBL5 | 20319 | -3.114 | -0.0432 | No | ||
| 166 | 5830411E10RIK | 20645 | -3.393 | -0.0513 | No | ||
| 167 | USP15 | 20781 | -3.542 | -0.0504 | No | ||
| 168 | 2310028O11RIK | 20949 | -3.750 | -0.0505 | No | ||
| 169 | HSPE1 | 21303 | -4.365 | -0.0580 | No | ||
| 170 | BCL2 | 21470 | -4.794 | -0.0560 | No | ||
| 171 | DUSP19 | 21534 | -5.009 | -0.0488 | No | ||
| 172 | PCBD2 | 21609 | -5.267 | -0.0416 | No | ||
| 173 | WASPIP | 21612 | -5.284 | -0.0311 | No | ||
| 174 | PAFAH1B1 | 21628 | -5.352 | -0.0210 | No | ||
| 175 | ZFP62 | 21653 | -5.453 | -0.0111 | No | ||
| 176 | MRPL9 | 21744 | -6.044 | -0.0031 | No | ||
| 177 | TIMM22 | 21764 | -6.171 | 0.0084 | No |