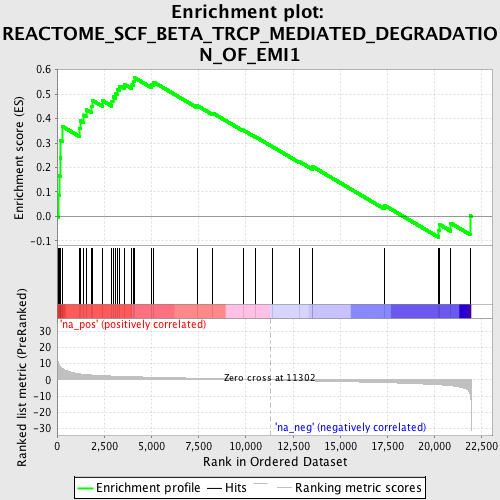
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set01_ATM_minus_versus_SCID |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_pos |
| GeneSet | REACTOME_SCF_BETA_TRCP_MEDIATED_DEGRADATION_OF_EMI1 |
| Enrichment Score (ES) | 0.56776184 |
| Normalized Enrichment Score (NES) | 1.972043 |
| Nominal p-value | 0.0017182131 |
| FDR q-value | 0.014055888 |
| FWER p-Value | 0.228 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | PSMD14 | 67 | 10.195 | 0.0880 | Yes | ||
| 2 | PSMC2 | 114 | 9.034 | 0.1666 | Yes | ||
| 3 | PSMC6 | 160 | 8.356 | 0.2391 | Yes | ||
| 4 | PSMD12 | 183 | 8.008 | 0.3096 | Yes | ||
| 5 | CUL1 | 279 | 6.999 | 0.3678 | Yes | ||
| 6 | PSMD5 | 1197 | 3.724 | 0.3592 | Yes | ||
| 7 | PSMB4 | 1225 | 3.687 | 0.3909 | Yes | ||
| 8 | PSMA4 | 1379 | 3.447 | 0.4147 | Yes | ||
| 9 | PSMA2 | 1560 | 3.252 | 0.4355 | Yes | ||
| 10 | PSMA6 | 1834 | 2.997 | 0.4498 | Yes | ||
| 11 | PSMD7 | 1879 | 2.960 | 0.4742 | Yes | ||
| 12 | PSMB1 | 2391 | 2.593 | 0.4741 | Yes | ||
| 13 | PSMD9 | 2904 | 2.340 | 0.4716 | Yes | ||
| 14 | PSMC5 | 2965 | 2.316 | 0.4895 | Yes | ||
| 15 | PSMA3 | 3100 | 2.259 | 0.5036 | Yes | ||
| 16 | PSMA1 | 3180 | 2.227 | 0.5199 | Yes | ||
| 17 | PSME3 | 3312 | 2.176 | 0.5333 | Yes | ||
| 18 | PSMD8 | 3556 | 2.078 | 0.5408 | Yes | ||
| 19 | PSMD6 | 3966 | 1.940 | 0.5395 | Yes | ||
| 20 | PSMC3 | 4064 | 1.905 | 0.5521 | Yes | ||
| 21 | PSMD4 | 4092 | 1.897 | 0.5678 | Yes | ||
| 22 | PSMC4 | 5016 | 1.600 | 0.5399 | No | ||
| 23 | PSMA7 | 5110 | 1.569 | 0.5497 | No | ||
| 24 | PSMB10 | 7426 | 0.999 | 0.4530 | No | ||
| 25 | PSMD3 | 8256 | 0.811 | 0.4224 | No | ||
| 26 | PSMC1 | 9853 | 0.423 | 0.3533 | No | ||
| 27 | PSMB3 | 10496 | 0.241 | 0.3262 | No | ||
| 28 | PSMB6 | 11428 | -0.044 | 0.2841 | No | ||
| 29 | PSME1 | 12817 | -0.453 | 0.2248 | No | ||
| 30 | PSMA5 | 13516 | -0.653 | 0.1988 | No | ||
| 31 | PSMB5 | 13523 | -0.657 | 0.2043 | No | ||
| 32 | PSMD2 | 17338 | -1.696 | 0.0454 | No | ||
| 33 | PSMB2 | 20232 | -3.048 | -0.0594 | No | ||
| 34 | PSMD11 | 20244 | -3.055 | -0.0326 | No | ||
| 35 | PSMD10 | 20860 | -3.644 | -0.0281 | No | ||
| 36 | PSMB9 | 21883 | -8.701 | 0.0029 | No |
