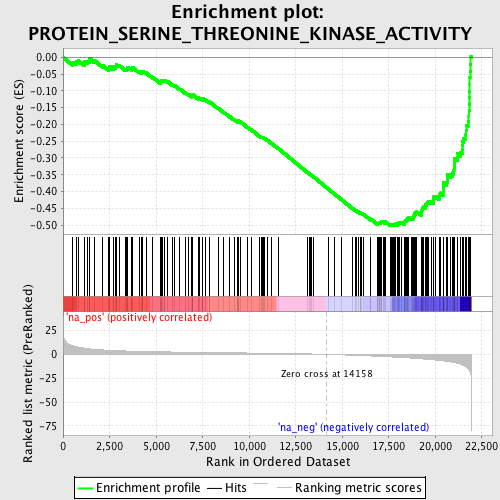
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set01_ATM_minus_versus_ATM_plus |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_neg |
| GeneSet | PROTEIN_SERINE_THREONINE_KINASE_ACTIVITY |
| Enrichment Score (ES) | -0.5027784 |
| Normalized Enrichment Score (NES) | -2.0930948 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.011538884 |
| FWER p-Value | 0.037 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | XRCC6BP1 | 519 | 8.900 | -0.0138 | No | ||
| 2 | CHEK2 | 707 | 7.951 | -0.0135 | No | ||
| 3 | CSNK2B | 803 | 7.574 | -0.0093 | No | ||
| 4 | RIPK4 | 1133 | 6.476 | -0.0172 | No | ||
| 5 | RIPK3 | 1159 | 6.396 | -0.0111 | No | ||
| 6 | PLK4 | 1306 | 6.023 | -0.0111 | No | ||
| 7 | CDC42BPA | 1415 | 5.784 | -0.0095 | No | ||
| 8 | MAPK1 | 1423 | 5.768 | -0.0033 | No | ||
| 9 | TSSK4 | 1673 | 5.287 | -0.0088 | No | ||
| 10 | TRIO | 2114 | 4.664 | -0.0238 | No | ||
| 11 | PBK | 2430 | 4.285 | -0.0335 | No | ||
| 12 | RPS6KA3 | 2494 | 4.216 | -0.0316 | No | ||
| 13 | PHKA1 | 2499 | 4.212 | -0.0271 | No | ||
| 14 | EIF2AK2 | 2700 | 4.017 | -0.0317 | No | ||
| 15 | MAPKAPK2 | 2720 | 3.998 | -0.0281 | No | ||
| 16 | NEK2 | 2823 | 3.930 | -0.0284 | No | ||
| 17 | TSSK6 | 2842 | 3.914 | -0.0248 | No | ||
| 18 | MAPK12 | 2869 | 3.894 | -0.0216 | No | ||
| 19 | ACVR1 | 3012 | 3.791 | -0.0239 | No | ||
| 20 | TTN | 3353 | 3.533 | -0.0355 | No | ||
| 21 | MAP3K6 | 3427 | 3.476 | -0.0350 | No | ||
| 22 | TTK | 3448 | 3.461 | -0.0320 | No | ||
| 23 | MAST1 | 3467 | 3.451 | -0.0290 | No | ||
| 24 | PLK1 | 3661 | 3.326 | -0.0341 | No | ||
| 25 | STK19 | 3697 | 3.311 | -0.0320 | No | ||
| 26 | LATS2 | 3716 | 3.300 | -0.0291 | No | ||
| 27 | VRK1 | 4077 | 3.102 | -0.0421 | No | ||
| 28 | UHMK1 | 4216 | 3.035 | -0.0451 | No | ||
| 29 | PRKCI | 4238 | 3.023 | -0.0426 | No | ||
| 30 | VRK2 | 4288 | 3.001 | -0.0415 | No | ||
| 31 | MYLK | 4466 | 2.925 | -0.0463 | No | ||
| 32 | EIF2AK4 | 4802 | 2.792 | -0.0586 | No | ||
| 33 | MAP3K9 | 5225 | 2.645 | -0.0750 | No | ||
| 34 | RPS6KA2 | 5248 | 2.639 | -0.0731 | No | ||
| 35 | RPS6KA1 | 5264 | 2.635 | -0.0708 | No | ||
| 36 | RIPK1 | 5284 | 2.628 | -0.0687 | No | ||
| 37 | BRSK1 | 5357 | 2.606 | -0.0691 | No | ||
| 38 | MAPK10 | 5422 | 2.586 | -0.0691 | No | ||
| 39 | LIMK1 | 5462 | 2.569 | -0.0680 | No | ||
| 40 | STK3 | 5627 | 2.519 | -0.0727 | No | ||
| 41 | IRAK3 | 5862 | 2.443 | -0.0807 | No | ||
| 42 | CDK3 | 5999 | 2.396 | -0.0843 | No | ||
| 43 | KALRN | 6274 | 2.311 | -0.0943 | No | ||
| 44 | MAPK4 | 6559 | 2.234 | -0.1048 | No | ||
| 45 | ACVRL1 | 6762 | 2.181 | -0.1116 | No | ||
| 46 | MAPK15 | 6907 | 2.138 | -0.1159 | No | ||
| 47 | HUNK | 6946 | 2.132 | -0.1152 | No | ||
| 48 | ACVR2A | 6956 | 2.130 | -0.1132 | No | ||
| 49 | MAP3K7 | 6975 | 2.124 | -0.1117 | No | ||
| 50 | SRPK1 | 7263 | 2.046 | -0.1225 | No | ||
| 51 | ACVR2B | 7271 | 2.044 | -0.1206 | No | ||
| 52 | RPS6KA5 | 7337 | 2.027 | -0.1213 | No | ||
| 53 | LATS1 | 7482 | 1.992 | -0.1257 | No | ||
| 54 | BRSK2 | 7492 | 1.989 | -0.1238 | No | ||
| 55 | BCKDK | 7507 | 1.985 | -0.1222 | No | ||
| 56 | WNK2 | 7648 | 1.947 | -0.1265 | No | ||
| 57 | IRAK1 | 7859 | 1.892 | -0.1340 | No | ||
| 58 | NEK4 | 7878 | 1.888 | -0.1327 | No | ||
| 59 | TSSK3 | 8327 | 1.782 | -0.1513 | No | ||
| 60 | MAP3K2 | 8636 | 1.709 | -0.1635 | No | ||
| 61 | NEK11 | 8953 | 1.637 | -0.1762 | No | ||
| 62 | WNK3 | 9218 | 1.571 | -0.1866 | No | ||
| 63 | ROCK2 | 9365 | 1.537 | -0.1916 | No | ||
| 64 | SGK2 | 9392 | 1.532 | -0.1910 | No | ||
| 65 | MARK1 | 9398 | 1.530 | -0.1895 | No | ||
| 66 | MAP3K4 | 9437 | 1.520 | -0.1896 | No | ||
| 67 | PRKD1 | 9525 | 1.498 | -0.1919 | No | ||
| 68 | BCR | 9930 | 1.403 | -0.2089 | No | ||
| 69 | WNK4 | 10123 | 1.354 | -0.2162 | No | ||
| 70 | PRKCZ | 10572 | 1.242 | -0.2354 | No | ||
| 71 | ACVR1C | 10669 | 1.222 | -0.2384 | No | ||
| 72 | DAPK2 | 10688 | 1.217 | -0.2379 | No | ||
| 73 | MAPK13 | 10765 | 1.197 | -0.2400 | No | ||
| 74 | CDK5 | 10811 | 1.185 | -0.2407 | No | ||
| 75 | CDK2 | 10991 | 1.139 | -0.2477 | No | ||
| 76 | STK36 | 11194 | 1.087 | -0.2558 | No | ||
| 77 | NEK6 | 11571 | 0.974 | -0.2719 | No | ||
| 78 | ERN2 | 13142 | 0.460 | -0.3436 | No | ||
| 79 | NUAK2 | 13262 | 0.412 | -0.3486 | No | ||
| 80 | TSSK2 | 13314 | 0.389 | -0.3505 | No | ||
| 81 | SNF1LK | 13371 | 0.369 | -0.3526 | No | ||
| 82 | PIM1 | 13451 | 0.338 | -0.3559 | No | ||
| 83 | PHKG2 | 14273 | -0.074 | -0.3935 | No | ||
| 84 | GTF2H1 | 14585 | -0.265 | -0.4075 | No | ||
| 85 | STK38L | 14967 | -0.508 | -0.4245 | No | ||
| 86 | RPS6KC1 | 15540 | -0.910 | -0.4497 | No | ||
| 87 | PLK3 | 15685 | -1.005 | -0.4552 | No | ||
| 88 | MAP4K3 | 15752 | -1.048 | -0.4571 | No | ||
| 89 | TESK1 | 15849 | -1.118 | -0.4602 | No | ||
| 90 | STK11 | 15968 | -1.215 | -0.4643 | No | ||
| 91 | CDK9 | 15989 | -1.229 | -0.4638 | No | ||
| 92 | CDC42BPB | 16031 | -1.265 | -0.4643 | No | ||
| 93 | RIPK2 | 16143 | -1.360 | -0.4679 | No | ||
| 94 | CPNE3 | 16523 | -1.676 | -0.4834 | No | ||
| 95 | TAF1 | 16532 | -1.684 | -0.4819 | No | ||
| 96 | CDKL1 | 16876 | -2.009 | -0.4954 | No | ||
| 97 | TNIK | 16901 | -2.033 | -0.4942 | No | ||
| 98 | PRKCH | 16959 | -2.083 | -0.4945 | No | ||
| 99 | DMPK | 16977 | -2.101 | -0.4929 | No | ||
| 100 | MAPK7 | 16999 | -2.122 | -0.4915 | No | ||
| 101 | AKT1 | 17066 | -2.193 | -0.4920 | No | ||
| 102 | DAPK3 | 17080 | -2.200 | -0.4901 | No | ||
| 103 | BRDT | 17102 | -2.222 | -0.4886 | No | ||
| 104 | MAPK9 | 17214 | -2.310 | -0.4911 | No | ||
| 105 | CDC42BPG | 17294 | -2.382 | -0.4921 | No | ||
| 106 | IRAK2 | 17297 | -2.389 | -0.4895 | No | ||
| 107 | CIT | 17582 | -2.678 | -0.4995 | No | ||
| 108 | CHUK | 17643 | -2.730 | -0.4992 | No | ||
| 109 | MARK4 | 17722 | -2.802 | -0.4996 | Yes | ||
| 110 | PAK1 | 17746 | -2.826 | -0.4975 | Yes | ||
| 111 | MAPK14 | 17811 | -2.903 | -0.4972 | Yes | ||
| 112 | MAP3K8 | 17860 | -2.954 | -0.4961 | Yes | ||
| 113 | RPS6KB2 | 17956 | -3.060 | -0.4970 | Yes | ||
| 114 | CDK7 | 18002 | -3.105 | -0.4956 | Yes | ||
| 115 | MAPK11 | 18040 | -3.143 | -0.4937 | Yes | ||
| 116 | MKNK2 | 18089 | -3.199 | -0.4923 | Yes | ||
| 117 | PHKA2 | 18186 | -3.318 | -0.4930 | Yes | ||
| 118 | EXOSC10 | 18318 | -3.438 | -0.4952 | Yes | ||
| 119 | CDK10 | 18341 | -3.463 | -0.4923 | Yes | ||
| 120 | MKNK1 | 18365 | -3.498 | -0.4894 | Yes | ||
| 121 | OXSR1 | 18380 | -3.517 | -0.4861 | Yes | ||
| 122 | STK16 | 18441 | -3.592 | -0.4848 | Yes | ||
| 123 | EIF2AK1 | 18481 | -3.633 | -0.4825 | Yes | ||
| 124 | ILK | 18512 | -3.670 | -0.4798 | Yes | ||
| 125 | CLK1 | 18579 | -3.733 | -0.4786 | Yes | ||
| 126 | GAK | 18704 | -3.882 | -0.4799 | Yes | ||
| 127 | MAP3K10 | 18760 | -3.957 | -0.4780 | Yes | ||
| 128 | ERN1 | 18848 | -4.074 | -0.4774 | Yes | ||
| 129 | ATM | 18851 | -4.077 | -0.4729 | Yes | ||
| 130 | PAK2 | 18865 | -4.102 | -0.4689 | Yes | ||
| 131 | CSNK1G2 | 18900 | -4.144 | -0.4658 | Yes | ||
| 132 | SGK3 | 18946 | -4.207 | -0.4631 | Yes | ||
| 133 | EIF2AK3 | 18978 | -4.241 | -0.4598 | Yes | ||
| 134 | BRD2 | 19233 | -4.628 | -0.4663 | Yes | ||
| 135 | TESK2 | 19259 | -4.659 | -0.4622 | Yes | ||
| 136 | PINK1 | 19279 | -4.705 | -0.4578 | Yes | ||
| 137 | PRKAG1 | 19286 | -4.710 | -0.4527 | Yes | ||
| 138 | MAP3K12 | 19326 | -4.778 | -0.4491 | Yes | ||
| 139 | MAP3K11 | 19341 | -4.796 | -0.4444 | Yes | ||
| 140 | PRKD3 | 19453 | -4.974 | -0.4439 | Yes | ||
| 141 | ULK1 | 19480 | -5.004 | -0.4395 | Yes | ||
| 142 | CSNK1D | 19547 | -5.118 | -0.4367 | Yes | ||
| 143 | CSNK1A1 | 19558 | -5.138 | -0.4314 | Yes | ||
| 144 | PRKX | 19622 | -5.230 | -0.4284 | Yes | ||
| 145 | CLK2 | 19812 | -5.514 | -0.4309 | Yes | ||
| 146 | PRKACB | 19908 | -5.677 | -0.4289 | Yes | ||
| 147 | ACVR1B | 19911 | -5.677 | -0.4226 | Yes | ||
| 148 | CSNK2A2 | 19914 | -5.695 | -0.4163 | Yes | ||
| 149 | ROCK1 | 20004 | -5.882 | -0.4138 | Yes | ||
| 150 | TLK1 | 20203 | -6.276 | -0.4158 | Yes | ||
| 151 | AKT2 | 20209 | -6.297 | -0.4089 | Yes | ||
| 152 | SNF1LK2 | 20268 | -6.428 | -0.4044 | Yes | ||
| 153 | LIMK2 | 20416 | -6.771 | -0.4035 | Yes | ||
| 154 | STK10 | 20418 | -6.774 | -0.3959 | Yes | ||
| 155 | MAP3K3 | 20422 | -6.791 | -0.3884 | Yes | ||
| 156 | PIM2 | 20446 | -6.848 | -0.3818 | Yes | ||
| 157 | MAP3K13 | 20449 | -6.853 | -0.3742 | Yes | ||
| 158 | MAPKAPK5 | 20596 | -7.204 | -0.3728 | Yes | ||
| 159 | DYRK1A | 20638 | -7.321 | -0.3664 | Yes | ||
| 160 | MINK1 | 20643 | -7.336 | -0.3584 | Yes | ||
| 161 | PRKCE | 20653 | -7.365 | -0.3505 | Yes | ||
| 162 | STK4 | 20820 | -7.881 | -0.3493 | Yes | ||
| 163 | CSNK2A1 | 20926 | -8.269 | -0.3448 | Yes | ||
| 164 | WNK1 | 20997 | -8.567 | -0.3384 | Yes | ||
| 165 | MAPK6 | 21006 | -8.592 | -0.3291 | Yes | ||
| 166 | MAP4K4 | 21016 | -8.644 | -0.3197 | Yes | ||
| 167 | TGFBR2 | 21043 | -8.737 | -0.3111 | Yes | ||
| 168 | TAOK3 | 21049 | -8.758 | -0.3015 | Yes | ||
| 169 | LMTK2 | 21182 | -9.262 | -0.2971 | Yes | ||
| 170 | MAPK8 | 21208 | -9.408 | -0.2877 | Yes | ||
| 171 | TGFBR1 | 21368 | -10.287 | -0.2834 | Yes | ||
| 172 | MAP4K5 | 21442 | -10.831 | -0.2746 | Yes | ||
| 173 | GSK3B | 21455 | -10.935 | -0.2629 | Yes | ||
| 174 | SRPK2 | 21476 | -11.046 | -0.2513 | Yes | ||
| 175 | SNRK | 21532 | -11.545 | -0.2409 | Yes | ||
| 176 | TLK2 | 21642 | -12.867 | -0.2314 | Yes | ||
| 177 | DYRK2 | 21648 | -12.985 | -0.2170 | Yes | ||
| 178 | HIPK3 | 21676 | -13.309 | -0.2033 | Yes | ||
| 179 | STK38 | 21758 | -14.684 | -0.1905 | Yes | ||
| 180 | CSNK1E | 21807 | -15.890 | -0.1748 | Yes | ||
| 181 | IKBKE | 21809 | -15.920 | -0.1570 | Yes | ||
| 182 | TAOK2 | 21830 | -16.810 | -0.1390 | Yes | ||
| 183 | MAP4K2 | 21842 | -17.169 | -0.1202 | Yes | ||
| 184 | CLK3 | 21849 | -17.494 | -0.1008 | Yes | ||
| 185 | MARK2 | 21858 | -17.748 | -0.0812 | Yes | ||
| 186 | STK17B | 21860 | -17.835 | -0.0612 | Yes | ||
| 187 | PRKCB1 | 21866 | -17.985 | -0.0412 | Yes | ||
| 188 | PDPK1 | 21879 | -18.696 | -0.0207 | Yes | ||
| 189 | NLK | 21913 | -21.118 | 0.0016 | Yes |