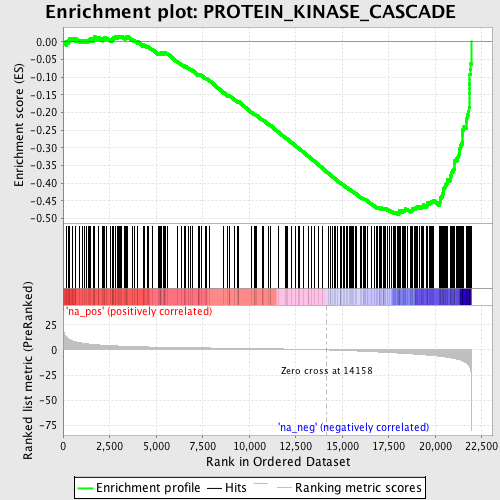
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set01_ATM_minus_versus_ATM_plus |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_neg |
| GeneSet | PROTEIN_KINASE_CASCADE |
| Enrichment Score (ES) | -0.48824304 |
| Normalized Enrichment Score (NES) | -2.134233 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.014972797 |
| FWER p-Value | 0.016 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | PRDX4 | 193 | 12.146 | 0.0008 | No | ||
| 2 | DUSP9 | 291 | 10.801 | 0.0050 | No | ||
| 3 | EEF1D | 360 | 10.178 | 0.0100 | No | ||
| 4 | TFG | 503 | 9.004 | 0.0107 | No | ||
| 5 | SLC35B2 | 689 | 8.017 | 0.0085 | No | ||
| 6 | TNFSF10 | 893 | 7.236 | 0.0050 | No | ||
| 7 | HMOX1 | 1027 | 6.810 | 0.0043 | No | ||
| 8 | ZDHHC17 | 1140 | 6.457 | 0.0043 | No | ||
| 9 | BST2 | 1274 | 6.105 | 0.0030 | No | ||
| 10 | DUSP22 | 1346 | 5.915 | 0.0045 | No | ||
| 11 | F2 | 1399 | 5.815 | 0.0067 | No | ||
| 12 | BLK | 1450 | 5.692 | 0.0090 | No | ||
| 13 | ITGB1BP1 | 1608 | 5.394 | 0.0061 | No | ||
| 14 | ECM1 | 1610 | 5.389 | 0.0103 | No | ||
| 15 | CCL2 | 1663 | 5.299 | 0.0122 | No | ||
| 16 | C5AR1 | 1709 | 5.233 | 0.0143 | No | ||
| 17 | VAPA | 1889 | 4.961 | 0.0100 | No | ||
| 18 | MAPK8IP1 | 1902 | 4.938 | 0.0134 | No | ||
| 19 | EDG2 | 2136 | 4.627 | 0.0064 | No | ||
| 20 | SOD1 | 2146 | 4.618 | 0.0096 | No | ||
| 21 | TGFA | 2218 | 4.525 | 0.0100 | No | ||
| 22 | ADRA2A | 2222 | 4.513 | 0.0135 | No | ||
| 23 | NRTN | 2352 | 4.360 | 0.0110 | No | ||
| 24 | ECT2 | 2560 | 4.143 | 0.0048 | No | ||
| 25 | RHOC | 2629 | 4.077 | 0.0049 | No | ||
| 26 | PLK2 | 2644 | 4.057 | 0.0075 | No | ||
| 27 | PROK2 | 2669 | 4.044 | 0.0096 | No | ||
| 28 | EDG5 | 2714 | 4.003 | 0.0108 | No | ||
| 29 | MAPKAPK2 | 2720 | 3.998 | 0.0138 | No | ||
| 30 | GOLT1B | 2794 | 3.957 | 0.0136 | No | ||
| 31 | RPL17 | 2811 | 3.941 | 0.0160 | No | ||
| 32 | FAF1 | 2945 | 3.834 | 0.0129 | No | ||
| 33 | GPR89A | 2971 | 3.820 | 0.0148 | No | ||
| 34 | NUP62 | 3002 | 3.797 | 0.0165 | No | ||
| 35 | IL31RA | 3090 | 3.740 | 0.0155 | No | ||
| 36 | SCG2 | 3146 | 3.697 | 0.0159 | No | ||
| 37 | ADRA2C | 3314 | 3.569 | 0.0110 | No | ||
| 38 | FPR1 | 3376 | 3.516 | 0.0110 | No | ||
| 39 | UBE2N | 3412 | 3.489 | 0.0122 | No | ||
| 40 | MAP3K6 | 3427 | 3.476 | 0.0143 | No | ||
| 41 | MAST1 | 3467 | 3.451 | 0.0153 | No | ||
| 42 | LATS2 | 3716 | 3.300 | 0.0065 | No | ||
| 43 | NF2 | 3843 | 3.233 | 0.0033 | No | ||
| 44 | PPP5C | 4007 | 3.142 | -0.0017 | No | ||
| 45 | LGALS9 | 4022 | 3.131 | 0.0002 | No | ||
| 46 | LGALS1 | 4293 | 2.998 | -0.0099 | No | ||
| 47 | NMI | 4302 | 2.995 | -0.0079 | No | ||
| 48 | GHRL | 4394 | 2.955 | -0.0097 | No | ||
| 49 | IRAK1BP1 | 4553 | 2.891 | -0.0147 | No | ||
| 50 | TNFRSF10B | 4565 | 2.887 | -0.0129 | No | ||
| 51 | PTPLAD1 | 4805 | 2.791 | -0.0217 | No | ||
| 52 | LTBR | 5104 | 2.685 | -0.0333 | No | ||
| 53 | ADAM9 | 5171 | 2.662 | -0.0342 | No | ||
| 54 | TMED4 | 5215 | 2.649 | -0.0340 | No | ||
| 55 | MAP3K9 | 5225 | 2.645 | -0.0323 | No | ||
| 56 | RPS6KA2 | 5248 | 2.639 | -0.0312 | No | ||
| 57 | RIPK1 | 5284 | 2.628 | -0.0307 | No | ||
| 58 | GSG2 | 5396 | 2.594 | -0.0338 | No | ||
| 59 | MAPK10 | 5422 | 2.586 | -0.0329 | No | ||
| 60 | GJA1 | 5432 | 2.583 | -0.0312 | No | ||
| 61 | ADORA2B | 5461 | 2.570 | -0.0305 | No | ||
| 62 | FGFR3 | 5506 | 2.555 | -0.0304 | No | ||
| 63 | STK3 | 5627 | 2.519 | -0.0340 | No | ||
| 64 | ADRB3 | 6121 | 2.360 | -0.0548 | No | ||
| 65 | ADRA1A | 6360 | 2.289 | -0.0639 | No | ||
| 66 | SOCS3 | 6520 | 2.246 | -0.0695 | No | ||
| 67 | IL12A | 6521 | 2.246 | -0.0677 | No | ||
| 68 | IL20 | 6572 | 2.232 | -0.0682 | No | ||
| 69 | FGF2 | 6738 | 2.186 | -0.0741 | No | ||
| 70 | FGFR1 | 6855 | 2.152 | -0.0777 | No | ||
| 71 | MAP3K7 | 6975 | 2.124 | -0.0815 | No | ||
| 72 | FASLG | 7259 | 2.047 | -0.0929 | No | ||
| 73 | SRPK1 | 7263 | 2.046 | -0.0914 | No | ||
| 74 | MAPK8IP2 | 7294 | 2.039 | -0.0911 | No | ||
| 75 | RPS6KA5 | 7337 | 2.027 | -0.0914 | No | ||
| 76 | STAMBP | 7439 | 2.002 | -0.0945 | No | ||
| 77 | WNK2 | 7648 | 1.947 | -0.1025 | No | ||
| 78 | MDFI | 7712 | 1.927 | -0.1039 | No | ||
| 79 | IRAK1 | 7859 | 1.892 | -0.1091 | No | ||
| 80 | MAP3K2 | 8636 | 1.709 | -0.1435 | No | ||
| 81 | DUSP8 | 8849 | 1.661 | -0.1520 | No | ||
| 82 | CD24 | 8918 | 1.644 | -0.1538 | No | ||
| 83 | TPD52L1 | 8920 | 1.643 | -0.1525 | No | ||
| 84 | NEK11 | 8953 | 1.637 | -0.1527 | No | ||
| 85 | WNK3 | 9218 | 1.571 | -0.1636 | No | ||
| 86 | SGK2 | 9392 | 1.532 | -0.1704 | No | ||
| 87 | MARK1 | 9398 | 1.530 | -0.1694 | No | ||
| 88 | TLR6 | 9407 | 1.528 | -0.1685 | No | ||
| 89 | MAP3K4 | 9437 | 1.520 | -0.1686 | No | ||
| 90 | WNK4 | 10123 | 1.354 | -0.1991 | No | ||
| 91 | LITAF | 10262 | 1.320 | -0.2044 | No | ||
| 92 | EDA2R | 10305 | 1.311 | -0.2053 | No | ||
| 93 | CHRNA7 | 10311 | 1.308 | -0.2045 | No | ||
| 94 | GNG3 | 10401 | 1.286 | -0.2076 | No | ||
| 95 | DAPK2 | 10688 | 1.217 | -0.2198 | No | ||
| 96 | CASP1 | 10705 | 1.212 | -0.2196 | No | ||
| 97 | MAPK13 | 10765 | 1.197 | -0.2213 | No | ||
| 98 | CARD10 | 11020 | 1.130 | -0.2321 | No | ||
| 99 | TRADD | 11123 | 1.103 | -0.2360 | No | ||
| 100 | TRIM13 | 11134 | 1.100 | -0.2355 | No | ||
| 101 | NEK6 | 11571 | 0.974 | -0.2549 | No | ||
| 102 | GRM4 | 11922 | 0.864 | -0.2703 | No | ||
| 103 | DUSP10 | 11980 | 0.847 | -0.2723 | No | ||
| 104 | CASP8 | 12035 | 0.830 | -0.2741 | No | ||
| 105 | EGF | 12295 | 0.756 | -0.2854 | No | ||
| 106 | TDGF1 | 12467 | 0.703 | -0.2927 | No | ||
| 107 | PLEKHG5 | 12625 | 0.650 | -0.2995 | No | ||
| 108 | NF1 | 12700 | 0.627 | -0.3024 | No | ||
| 109 | CD81 | 12940 | 0.537 | -0.3130 | No | ||
| 110 | RGS4 | 13168 | 0.450 | -0.3231 | No | ||
| 111 | ADRA1B | 13187 | 0.441 | -0.3235 | No | ||
| 112 | SNF1LK | 13371 | 0.369 | -0.3317 | No | ||
| 113 | AKAP11 | 13528 | 0.305 | -0.3386 | No | ||
| 114 | GADD45G | 13718 | 0.218 | -0.3472 | No | ||
| 115 | AMBP | 13939 | 0.112 | -0.3572 | No | ||
| 116 | MYD88 | 14281 | -0.077 | -0.3729 | No | ||
| 117 | GPS2 | 14368 | -0.134 | -0.3767 | No | ||
| 118 | CARTPT | 14372 | -0.135 | -0.3768 | No | ||
| 119 | ATP6AP2 | 14483 | -0.212 | -0.3817 | No | ||
| 120 | PLCE1 | 14566 | -0.256 | -0.3853 | No | ||
| 121 | HTR2B | 14615 | -0.277 | -0.3872 | No | ||
| 122 | DUSP4 | 14744 | -0.376 | -0.3928 | No | ||
| 123 | SECTM1 | 14880 | -0.466 | -0.3987 | No | ||
| 124 | ICK | 14903 | -0.477 | -0.3993 | No | ||
| 125 | REL | 14913 | -0.486 | -0.3994 | No | ||
| 126 | STK38L | 14967 | -0.508 | -0.4014 | No | ||
| 127 | TSPAN6 | 15046 | -0.572 | -0.4045 | No | ||
| 128 | PIK3CB | 15122 | -0.630 | -0.4075 | No | ||
| 129 | SRC | 15241 | -0.705 | -0.4124 | No | ||
| 130 | CXXC5 | 15268 | -0.719 | -0.4130 | No | ||
| 131 | TMEM101 | 15378 | -0.792 | -0.4174 | No | ||
| 132 | CD40 | 15416 | -0.823 | -0.4184 | No | ||
| 133 | CCR2 | 15486 | -0.869 | -0.4209 | No | ||
| 134 | TRAF7 | 15556 | -0.921 | -0.4234 | No | ||
| 135 | GPS1 | 15608 | -0.951 | -0.4249 | No | ||
| 136 | MBIP | 15706 | -1.018 | -0.4286 | No | ||
| 137 | MAP4K3 | 15752 | -1.048 | -0.4298 | No | ||
| 138 | ADRB2 | 15984 | -1.226 | -0.4395 | No | ||
| 139 | ZDHHC13 | 16051 | -1.284 | -0.4415 | No | ||
| 140 | CAMKK2 | 16123 | -1.344 | -0.4437 | No | ||
| 141 | RIPK2 | 16143 | -1.360 | -0.4435 | No | ||
| 142 | MIB2 | 16189 | -1.401 | -0.4445 | No | ||
| 143 | DAPK1 | 16250 | -1.448 | -0.4461 | No | ||
| 144 | DAXX | 16329 | -1.510 | -0.4485 | No | ||
| 145 | F2R | 16595 | -1.749 | -0.4593 | No | ||
| 146 | SHC1 | 16723 | -1.857 | -0.4637 | No | ||
| 147 | RPS6KA4 | 16863 | -1.997 | -0.4685 | No | ||
| 148 | TNIK | 16901 | -2.033 | -0.4685 | No | ||
| 149 | TRAF6 | 16980 | -2.105 | -0.4705 | No | ||
| 150 | HCLS1 | 17013 | -2.142 | -0.4702 | No | ||
| 151 | RHOA | 17075 | -2.199 | -0.4713 | No | ||
| 152 | DAPK3 | 17080 | -2.200 | -0.4697 | No | ||
| 153 | MAP3K7IP1 | 17192 | -2.295 | -0.4730 | No | ||
| 154 | MAPK9 | 17214 | -2.310 | -0.4721 | No | ||
| 155 | TNFSF14 | 17226 | -2.321 | -0.4708 | No | ||
| 156 | ERC1 | 17266 | -2.356 | -0.4707 | No | ||
| 157 | IRAK2 | 17297 | -2.389 | -0.4701 | No | ||
| 158 | HGS | 17453 | -2.539 | -0.4753 | No | ||
| 159 | SPRED1 | 17558 | -2.657 | -0.4779 | No | ||
| 160 | CHUK | 17643 | -2.730 | -0.4796 | No | ||
| 161 | PAK1 | 17746 | -2.826 | -0.4821 | No | ||
| 162 | MAPK14 | 17811 | -2.903 | -0.4827 | No | ||
| 163 | STAT4 | 17839 | -2.936 | -0.4816 | No | ||
| 164 | FKBP1A | 17973 | -3.076 | -0.4852 | No | ||
| 165 | OTUD7B | 18039 | -3.141 | -0.4857 | Yes | ||
| 166 | MAPK11 | 18040 | -3.143 | -0.4832 | Yes | ||
| 167 | TMEM9B | 18073 | -3.182 | -0.4821 | Yes | ||
| 168 | SOCS2 | 18085 | -3.196 | -0.4801 | Yes | ||
| 169 | MKNK2 | 18089 | -3.199 | -0.4777 | Yes | ||
| 170 | RGS3 | 18102 | -3.221 | -0.4757 | Yes | ||
| 171 | TICAM1 | 18233 | -3.360 | -0.4790 | Yes | ||
| 172 | FADD | 18270 | -3.402 | -0.4779 | Yes | ||
| 173 | BCL3 | 18305 | -3.430 | -0.4767 | Yes | ||
| 174 | MKNK1 | 18365 | -3.498 | -0.4766 | Yes | ||
| 175 | OXSR1 | 18380 | -3.517 | -0.4745 | Yes | ||
| 176 | IKBKG | 18397 | -3.530 | -0.4724 | Yes | ||
| 177 | DBNL | 18517 | -3.675 | -0.4749 | Yes | ||
| 178 | FYB | 18658 | -3.819 | -0.4783 | Yes | ||
| 179 | DUSP6 | 18680 | -3.854 | -0.4762 | Yes | ||
| 180 | TBK1 | 18737 | -3.926 | -0.4757 | Yes | ||
| 181 | SOCS6 | 18758 | -3.954 | -0.4734 | Yes | ||
| 182 | MAP3K10 | 18760 | -3.957 | -0.4703 | Yes | ||
| 183 | CXCR4 | 18871 | -4.113 | -0.4721 | Yes | ||
| 184 | GPR177 | 18914 | -4.167 | -0.4707 | Yes | ||
| 185 | IFNAR2 | 18945 | -4.206 | -0.4687 | Yes | ||
| 186 | HIPK2 | 18997 | -4.265 | -0.4676 | Yes | ||
| 187 | TRIP6 | 19029 | -4.323 | -0.4656 | Yes | ||
| 188 | TRAF3IP2 | 19126 | -4.464 | -0.4665 | Yes | ||
| 189 | PTEN | 19245 | -4.642 | -0.4682 | Yes | ||
| 190 | PINK1 | 19279 | -4.705 | -0.4660 | Yes | ||
| 191 | MAP3K12 | 19326 | -4.778 | -0.4642 | Yes | ||
| 192 | MAP3K11 | 19341 | -4.796 | -0.4611 | Yes | ||
| 193 | SH2D3C | 19505 | -5.042 | -0.4645 | Yes | ||
| 194 | SCOTIN | 19554 | -5.132 | -0.4626 | Yes | ||
| 195 | CANT1 | 19559 | -5.139 | -0.4587 | Yes | ||
| 196 | BIRC2 | 19566 | -5.145 | -0.4549 | Yes | ||
| 197 | STAT5A | 19672 | -5.285 | -0.4555 | Yes | ||
| 198 | CLCF1 | 19735 | -5.380 | -0.4540 | Yes | ||
| 199 | NDFIP1 | 19774 | -5.439 | -0.4515 | Yes | ||
| 200 | MDFIC | 19856 | -5.585 | -0.4507 | Yes | ||
| 201 | LAX1 | 19906 | -5.674 | -0.4484 | Yes | ||
| 202 | CC2D1A | 20201 | -6.272 | -0.4570 | Yes | ||
| 203 | BCL10 | 20231 | -6.352 | -0.4532 | Yes | ||
| 204 | MAP2K7 | 20261 | -6.417 | -0.4494 | Yes | ||
| 205 | SNF1LK2 | 20268 | -6.428 | -0.4446 | Yes | ||
| 206 | MAP3K7IP2 | 20281 | -6.446 | -0.4400 | Yes | ||
| 207 | ATP2C1 | 20325 | -6.547 | -0.4367 | Yes | ||
| 208 | PIAS1 | 20382 | -6.675 | -0.4340 | Yes | ||
| 209 | NDFIP2 | 20391 | -6.700 | -0.4290 | Yes | ||
| 210 | MAP3K3 | 20422 | -6.791 | -0.4249 | Yes | ||
| 211 | MAP3K13 | 20449 | -6.853 | -0.4206 | Yes | ||
| 212 | PKN1 | 20455 | -6.883 | -0.4154 | Yes | ||
| 213 | STAT2 | 20472 | -6.911 | -0.4106 | Yes | ||
| 214 | EDARADD | 20526 | -7.013 | -0.4074 | Yes | ||
| 215 | CRKL | 20555 | -7.087 | -0.4030 | Yes | ||
| 216 | SPRED2 | 20599 | -7.209 | -0.3992 | Yes | ||
| 217 | RHOH | 20639 | -7.326 | -0.3952 | Yes | ||
| 218 | MINK1 | 20643 | -7.336 | -0.3894 | Yes | ||
| 219 | STAT3 | 20789 | -7.787 | -0.3899 | Yes | ||
| 220 | STK4 | 20820 | -7.881 | -0.3850 | Yes | ||
| 221 | SLC20A1 | 20830 | -7.912 | -0.3791 | Yes | ||
| 222 | PPM1A | 20886 | -8.120 | -0.3751 | Yes | ||
| 223 | FLNA | 20889 | -8.130 | -0.3687 | Yes | ||
| 224 | DUSP16 | 20921 | -8.252 | -0.3635 | Yes | ||
| 225 | WNK1 | 20997 | -8.567 | -0.3601 | Yes | ||
| 226 | MAP4K4 | 21016 | -8.644 | -0.3540 | Yes | ||
| 227 | TRAF2 | 21018 | -8.648 | -0.3472 | Yes | ||
| 228 | RELA | 21022 | -8.671 | -0.3404 | Yes | ||
| 229 | TAOK3 | 21049 | -8.758 | -0.3346 | Yes | ||
| 230 | DUSP2 | 21113 | -8.980 | -0.3303 | Yes | ||
| 231 | MAPK8 | 21208 | -9.408 | -0.3271 | Yes | ||
| 232 | SLC44A2 | 21236 | -9.565 | -0.3207 | Yes | ||
| 233 | MADD | 21274 | -9.747 | -0.3146 | Yes | ||
| 234 | FYN | 21301 | -9.911 | -0.3079 | Yes | ||
| 235 | SQSTM1 | 21318 | -9.993 | -0.3006 | Yes | ||
| 236 | SOCS1 | 21356 | -10.225 | -0.2941 | Yes | ||
| 237 | GADD45B | 21406 | -10.572 | -0.2879 | Yes | ||
| 238 | MAP4K5 | 21442 | -10.831 | -0.2809 | Yes | ||
| 239 | CFLAR | 21465 | -10.989 | -0.2731 | Yes | ||
| 240 | MAP2K4 | 21466 | -10.991 | -0.2643 | Yes | ||
| 241 | SRPK2 | 21476 | -11.046 | -0.2559 | Yes | ||
| 242 | TNFAIP3 | 21482 | -11.148 | -0.2472 | Yes | ||
| 243 | CCM2 | 21538 | -11.589 | -0.2405 | Yes | ||
| 244 | STAT5B | 21655 | -13.076 | -0.2353 | Yes | ||
| 245 | STAT1 | 21656 | -13.091 | -0.2249 | Yes | ||
| 246 | TEC | 21697 | -13.757 | -0.2157 | Yes | ||
| 247 | TNFRSF1A | 21703 | -13.816 | -0.2049 | Yes | ||
| 248 | STK38 | 21758 | -14.684 | -0.1956 | Yes | ||
| 249 | IKBKE | 21809 | -15.920 | -0.1852 | Yes | ||
| 250 | MIER1 | 21829 | -16.787 | -0.1726 | Yes | ||
| 251 | TAOK2 | 21830 | -16.810 | -0.1592 | Yes | ||
| 252 | IFNAR1 | 21831 | -16.830 | -0.1457 | Yes | ||
| 253 | MAPK8IP3 | 21835 | -16.890 | -0.1324 | Yes | ||
| 254 | MAP4K2 | 21842 | -17.169 | -0.1189 | Yes | ||
| 255 | MARK2 | 21858 | -17.748 | -0.1054 | Yes | ||
| 256 | STK17B | 21860 | -17.835 | -0.0912 | Yes | ||
| 257 | MALT1 | 21871 | -18.066 | -0.0772 | Yes | ||
| 258 | NLK | 21913 | -21.118 | -0.0622 | Yes | ||
| 259 | FGF13 | 21947 | -79.619 | 0.0000 | Yes |