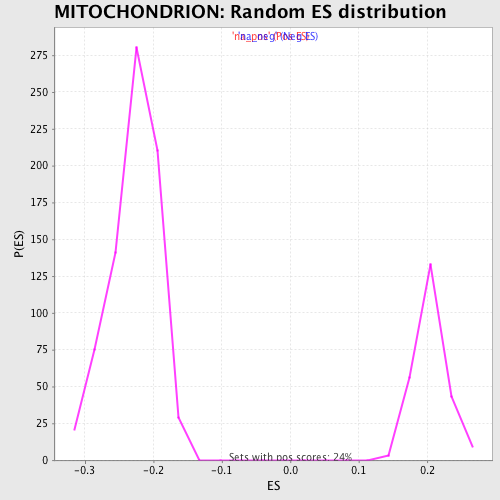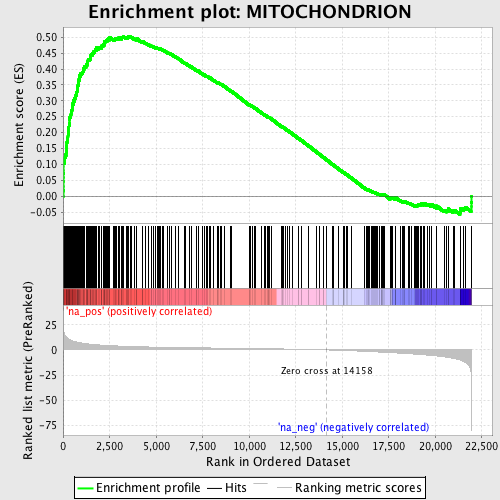
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set01_ATM_minus_versus_ATM_plus |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_pos |
| GeneSet | MITOCHONDRION |
| Enrichment Score (ES) | 0.50341964 |
| Normalized Enrichment Score (NES) | 2.4660454 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.0 |
| FWER p-Value | 0.0 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | MRPS10 | 4 | 25.608 | 0.0169 | Yes | ||
| 2 | HSPD1 | 14 | 22.156 | 0.0312 | Yes | ||
| 3 | ATP5G1 | 16 | 21.522 | 0.0455 | Yes | ||
| 4 | MECR | 21 | 19.891 | 0.0585 | Yes | ||
| 5 | SOD2 | 24 | 19.385 | 0.0713 | Yes | ||
| 6 | CYCS | 42 | 17.128 | 0.0819 | Yes | ||
| 7 | TIMM17A | 44 | 16.991 | 0.0932 | Yes | ||
| 8 | GOT2 | 45 | 16.934 | 0.1044 | Yes | ||
| 9 | PHB2 | 65 | 15.962 | 0.1142 | Yes | ||
| 10 | IMMT | 98 | 14.631 | 0.1224 | Yes | ||
| 11 | PHB | 132 | 13.657 | 0.1300 | Yes | ||
| 12 | TIMM50 | 161 | 12.723 | 0.1372 | Yes | ||
| 13 | TSFM | 171 | 12.539 | 0.1451 | Yes | ||
| 14 | HADH | 179 | 12.387 | 0.1530 | Yes | ||
| 15 | TXNRD2 | 195 | 12.115 | 0.1604 | Yes | ||
| 16 | CPT1A | 202 | 11.926 | 0.1681 | Yes | ||
| 17 | VDAC2 | 214 | 11.747 | 0.1754 | Yes | ||
| 18 | GRPEL1 | 220 | 11.637 | 0.1829 | Yes | ||
| 19 | BID | 260 | 11.198 | 0.1885 | Yes | ||
| 20 | MRPS36 | 263 | 11.174 | 0.1959 | Yes | ||
| 21 | SHMT2 | 279 | 10.992 | 0.2025 | Yes | ||
| 22 | NDUFA8 | 288 | 10.858 | 0.2093 | Yes | ||
| 23 | HSPE1 | 301 | 10.746 | 0.2159 | Yes | ||
| 24 | MRPL12 | 316 | 10.627 | 0.2224 | Yes | ||
| 25 | ABCE1 | 328 | 10.524 | 0.2289 | Yes | ||
| 26 | VDAC1 | 339 | 10.409 | 0.2353 | Yes | ||
| 27 | MTHFD1 | 340 | 10.406 | 0.2422 | Yes | ||
| 28 | SLC25A4 | 361 | 10.176 | 0.2481 | Yes | ||
| 29 | ETFA | 385 | 9.964 | 0.2537 | Yes | ||
| 30 | MRPS11 | 421 | 9.615 | 0.2584 | Yes | ||
| 31 | NDUFV1 | 429 | 9.497 | 0.2644 | Yes | ||
| 32 | NDUFAB1 | 465 | 9.257 | 0.2690 | Yes | ||
| 33 | SDHD | 480 | 9.166 | 0.2744 | Yes | ||
| 34 | TIMM9 | 489 | 9.084 | 0.2801 | Yes | ||
| 35 | VDAC3 | 516 | 8.932 | 0.2848 | Yes | ||
| 36 | MRPL40 | 524 | 8.847 | 0.2904 | Yes | ||
| 37 | TIMM10 | 566 | 8.600 | 0.2942 | Yes | ||
| 38 | MRPS18A | 567 | 8.597 | 0.3000 | Yes | ||
| 39 | SLC25A3 | 603 | 8.417 | 0.3039 | Yes | ||
| 40 | SDHB | 619 | 8.329 | 0.3088 | Yes | ||
| 41 | MRPS12 | 666 | 8.123 | 0.3121 | Yes | ||
| 42 | FDXR | 684 | 8.052 | 0.3166 | Yes | ||
| 43 | MRPS22 | 705 | 7.954 | 0.3210 | Yes | ||
| 44 | DECR1 | 713 | 7.936 | 0.3260 | Yes | ||
| 45 | MRPS35 | 746 | 7.786 | 0.3297 | Yes | ||
| 46 | OAT | 754 | 7.735 | 0.3345 | Yes | ||
| 47 | CYC1 | 767 | 7.693 | 0.3391 | Yes | ||
| 48 | MRPS18C | 783 | 7.653 | 0.3435 | Yes | ||
| 49 | COQ7 | 795 | 7.620 | 0.3480 | Yes | ||
| 50 | MRPL51 | 811 | 7.526 | 0.3523 | Yes | ||
| 51 | TRNT1 | 828 | 7.481 | 0.3566 | Yes | ||
| 52 | TIMM8B | 832 | 7.464 | 0.3614 | Yes | ||
| 53 | NDUFA11 | 848 | 7.395 | 0.3656 | Yes | ||
| 54 | PCCB | 871 | 7.322 | 0.3695 | Yes | ||
| 55 | DUT | 879 | 7.294 | 0.3740 | Yes | ||
| 56 | MIPEP | 886 | 7.270 | 0.3786 | Yes | ||
| 57 | MRPS28 | 930 | 7.107 | 0.3813 | Yes | ||
| 58 | MRPS15 | 953 | 7.020 | 0.3850 | Yes | ||
| 59 | OPA1 | 1003 | 6.867 | 0.3873 | Yes | ||
| 60 | ABCF2 | 1043 | 6.750 | 0.3900 | Yes | ||
| 61 | GSTZ1 | 1068 | 6.667 | 0.3933 | Yes | ||
| 62 | ATP5J | 1098 | 6.550 | 0.3963 | Yes | ||
| 63 | DLD | 1102 | 6.542 | 0.4005 | Yes | ||
| 64 | CHCHD3 | 1126 | 6.486 | 0.4038 | Yes | ||
| 65 | NDUFS8 | 1146 | 6.442 | 0.4072 | Yes | ||
| 66 | MRPL55 | 1231 | 6.214 | 0.4074 | Yes | ||
| 67 | NDUFA9 | 1233 | 6.204 | 0.4115 | Yes | ||
| 68 | GPX4 | 1265 | 6.125 | 0.4142 | Yes | ||
| 69 | COQ6 | 1302 | 6.035 | 0.4165 | Yes | ||
| 70 | TOMM22 | 1315 | 6.000 | 0.4200 | Yes | ||
| 71 | UQCRC1 | 1327 | 5.967 | 0.4234 | Yes | ||
| 72 | CS | 1336 | 5.959 | 0.4270 | Yes | ||
| 73 | NDUFS3 | 1351 | 5.899 | 0.4303 | Yes | ||
| 74 | ABCB7 | 1428 | 5.759 | 0.4306 | Yes | ||
| 75 | PECI | 1455 | 5.683 | 0.4332 | Yes | ||
| 76 | LRPPRC | 1473 | 5.634 | 0.4362 | Yes | ||
| 77 | OXA1L | 1476 | 5.629 | 0.4398 | Yes | ||
| 78 | PDK3 | 1488 | 5.612 | 0.4430 | Yes | ||
| 79 | ACP6 | 1504 | 5.590 | 0.4461 | Yes | ||
| 80 | COX4NB | 1560 | 5.478 | 0.4472 | Yes | ||
| 81 | ABCB6 | 1580 | 5.452 | 0.4499 | Yes | ||
| 82 | TBRG4 | 1629 | 5.351 | 0.4513 | Yes | ||
| 83 | ATP5F1 | 1653 | 5.316 | 0.4537 | Yes | ||
| 84 | NT5M | 1699 | 5.249 | 0.4551 | Yes | ||
| 85 | TMEM126A | 1713 | 5.227 | 0.4580 | Yes | ||
| 86 | NME4 | 1724 | 5.209 | 0.4610 | Yes | ||
| 87 | MRPS31 | 1790 | 5.089 | 0.4614 | Yes | ||
| 88 | MTCH1 | 1794 | 5.083 | 0.4647 | Yes | ||
| 89 | MTHFD2 | 1800 | 5.075 | 0.4678 | Yes | ||
| 90 | AFG3L2 | 1913 | 4.924 | 0.4659 | Yes | ||
| 91 | TIMM44 | 1946 | 4.883 | 0.4677 | Yes | ||
| 92 | FXN | 2047 | 4.745 | 0.4662 | Yes | ||
| 93 | ABCB8 | 2055 | 4.740 | 0.4690 | Yes | ||
| 94 | ATP5A1 | 2058 | 4.738 | 0.4721 | Yes | ||
| 95 | ECSIT | 2087 | 4.712 | 0.4739 | Yes | ||
| 96 | SOD1 | 2146 | 4.618 | 0.4743 | Yes | ||
| 97 | DHRS4 | 2170 | 4.571 | 0.4763 | Yes | ||
| 98 | BAX | 2201 | 4.543 | 0.4780 | Yes | ||
| 99 | ATP5O | 2209 | 4.538 | 0.4807 | Yes | ||
| 100 | MRPL23 | 2213 | 4.530 | 0.4835 | Yes | ||
| 101 | SLC25A1 | 2216 | 4.528 | 0.4865 | Yes | ||
| 102 | ACADS | 2254 | 4.466 | 0.4877 | Yes | ||
| 103 | ATP5E | 2306 | 4.413 | 0.4883 | Yes | ||
| 104 | MRPL32 | 2334 | 4.378 | 0.4900 | Yes | ||
| 105 | AK3 | 2380 | 4.328 | 0.4908 | Yes | ||
| 106 | SUCLA2 | 2400 | 4.309 | 0.4928 | Yes | ||
| 107 | TIMM23 | 2418 | 4.296 | 0.4948 | Yes | ||
| 108 | ESR2 | 2501 | 4.212 | 0.4938 | Yes | ||
| 109 | ECHS1 | 2508 | 4.202 | 0.4964 | Yes | ||
| 110 | IDH3B | 2511 | 4.199 | 0.4991 | Yes | ||
| 111 | PTGES2 | 2697 | 4.020 | 0.4932 | Yes | ||
| 112 | ATP5G3 | 2767 | 3.974 | 0.4926 | Yes | ||
| 113 | ACN9 | 2788 | 3.961 | 0.4944 | Yes | ||
| 114 | ATP5D | 2793 | 3.957 | 0.4968 | Yes | ||
| 115 | TP53 | 2880 | 3.885 | 0.4954 | Yes | ||
| 116 | FPGS | 2957 | 3.828 | 0.4945 | Yes | ||
| 117 | TIMM13 | 2968 | 3.821 | 0.4965 | Yes | ||
| 118 | TUFM | 2998 | 3.802 | 0.4977 | Yes | ||
| 119 | SLC25A15 | 3014 | 3.790 | 0.4996 | Yes | ||
| 120 | SUPV3L1 | 3135 | 3.706 | 0.4965 | Yes | ||
| 121 | MSTO1 | 3139 | 3.704 | 0.4988 | Yes | ||
| 122 | PITRM1 | 3162 | 3.688 | 0.5002 | Yes | ||
| 123 | AGPAT5 | 3215 | 3.643 | 0.5003 | Yes | ||
| 124 | OXSM | 3249 | 3.616 | 0.5011 | Yes | ||
| 125 | DNAJC19 | 3390 | 3.500 | 0.4970 | Yes | ||
| 126 | TRMU | 3395 | 3.498 | 0.4992 | Yes | ||
| 127 | BCS1L | 3457 | 3.457 | 0.4986 | Yes | ||
| 128 | HTRA2 | 3480 | 3.446 | 0.4999 | Yes | ||
| 129 | HSPA1B | 3500 | 3.436 | 0.5013 | Yes | ||
| 130 | NDUFB6 | 3505 | 3.432 | 0.5034 | Yes | ||
| 131 | ATP5G2 | 3603 | 3.359 | 0.5012 | No | ||
| 132 | SUCLG1 | 3674 | 3.321 | 0.5002 | No | ||
| 133 | SLC25A13 | 3857 | 3.225 | 0.4939 | No | ||
| 134 | ATP5C1 | 3949 | 3.174 | 0.4918 | No | ||
| 135 | ATP5B | 3951 | 3.174 | 0.4939 | No | ||
| 136 | GLUD1 | 3965 | 3.164 | 0.4954 | No | ||
| 137 | MRPS16 | 4239 | 3.023 | 0.4848 | No | ||
| 138 | UQCRB | 4258 | 3.014 | 0.4859 | No | ||
| 139 | MTX2 | 4291 | 3.000 | 0.4865 | No | ||
| 140 | PCK2 | 4449 | 2.932 | 0.4812 | No | ||
| 141 | NNT | 4589 | 2.880 | 0.4767 | No | ||
| 142 | GLS2 | 4728 | 2.824 | 0.4722 | No | ||
| 143 | DNAJA3 | 4764 | 2.809 | 0.4724 | No | ||
| 144 | ACOT2 | 4872 | 2.766 | 0.4693 | No | ||
| 145 | SARDH | 4955 | 2.736 | 0.4674 | No | ||
| 146 | MRPL41 | 4974 | 2.731 | 0.4683 | No | ||
| 147 | ME3 | 5088 | 2.690 | 0.4649 | No | ||
| 148 | COX15 | 5112 | 2.683 | 0.4656 | No | ||
| 149 | NDUFS7 | 5194 | 2.655 | 0.4637 | No | ||
| 150 | NDUFA1 | 5233 | 2.644 | 0.4637 | No | ||
| 151 | COX8C | 5364 | 2.603 | 0.4594 | No | ||
| 152 | ETFB | 5412 | 2.587 | 0.4589 | No | ||
| 153 | PTRF | 5607 | 2.523 | 0.4517 | No | ||
| 154 | NDUFA6 | 5703 | 2.494 | 0.4489 | No | ||
| 155 | HINT2 | 5738 | 2.483 | 0.4490 | No | ||
| 156 | SMCP | 5830 | 2.454 | 0.4464 | No | ||
| 157 | SDHC | 6017 | 2.390 | 0.4394 | No | ||
| 158 | MUT | 6185 | 2.340 | 0.4333 | No | ||
| 159 | UCP3 | 6536 | 2.242 | 0.4186 | No | ||
| 160 | GHITM | 6563 | 2.233 | 0.4189 | No | ||
| 161 | RAB11FIP5 | 6791 | 2.174 | 0.4099 | No | ||
| 162 | BDH2 | 6886 | 2.144 | 0.4069 | No | ||
| 163 | L2HGDH | 7157 | 2.075 | 0.3959 | No | ||
| 164 | GLRX5 | 7184 | 2.069 | 0.3960 | No | ||
| 165 | PSEN1 | 7252 | 2.051 | 0.3943 | No | ||
| 166 | BCKDK | 7507 | 1.985 | 0.3839 | No | ||
| 167 | PPOX | 7570 | 1.970 | 0.3823 | No | ||
| 168 | SARS2 | 7706 | 1.928 | 0.3774 | No | ||
| 169 | HAX1 | 7738 | 1.920 | 0.3772 | No | ||
| 170 | LETM1 | 7888 | 1.885 | 0.3716 | No | ||
| 171 | AASS | 7906 | 1.881 | 0.3721 | No | ||
| 172 | DMGDH | 8077 | 1.842 | 0.3654 | No | ||
| 173 | BAK1 | 8284 | 1.793 | 0.3571 | No | ||
| 174 | NDUFS2 | 8317 | 1.784 | 0.3568 | No | ||
| 175 | NDUFS5 | 8356 | 1.779 | 0.3563 | No | ||
| 176 | TMEM143 | 8477 | 1.748 | 0.3519 | No | ||
| 177 | MAOB | 8508 | 1.742 | 0.3517 | No | ||
| 178 | UQCRH | 8656 | 1.702 | 0.3460 | No | ||
| 179 | ARG2 | 9019 | 1.621 | 0.3304 | No | ||
| 180 | FOXRED1 | 9059 | 1.611 | 0.3296 | No | ||
| 181 | HCCS | 10013 | 1.379 | 0.2865 | No | ||
| 182 | BCL2L10 | 10016 | 1.379 | 0.2874 | No | ||
| 183 | TFB2M | 10069 | 1.366 | 0.2859 | No | ||
| 184 | HMGCS2 | 10168 | 1.343 | 0.2822 | No | ||
| 185 | TRIAP1 | 10296 | 1.313 | 0.2772 | No | ||
| 186 | LDHD | 10326 | 1.306 | 0.2768 | No | ||
| 187 | GLYAT | 10682 | 1.218 | 0.2612 | No | ||
| 188 | HSD3B1 | 10809 | 1.185 | 0.2562 | No | ||
| 189 | CENTA2 | 10854 | 1.174 | 0.2549 | No | ||
| 190 | BNIP3L | 10963 | 1.146 | 0.2507 | No | ||
| 191 | SCO1 | 11024 | 1.128 | 0.2487 | No | ||
| 192 | CKMT2 | 11055 | 1.120 | 0.2480 | No | ||
| 193 | PET112L | 11092 | 1.111 | 0.2471 | No | ||
| 194 | MSRB3 | 11202 | 1.084 | 0.2428 | No | ||
| 195 | GLRX2 | 11722 | 0.927 | 0.2194 | No | ||
| 196 | HADHB | 11770 | 0.914 | 0.2179 | No | ||
| 197 | MRPL10 | 11785 | 0.910 | 0.2178 | No | ||
| 198 | NDUFS1 | 11834 | 0.895 | 0.2162 | No | ||
| 199 | CPT1B | 11941 | 0.857 | 0.2119 | No | ||
| 200 | CASP8 | 12035 | 0.830 | 0.2081 | No | ||
| 201 | MRPS24 | 12152 | 0.797 | 0.2033 | No | ||
| 202 | GCDH | 12329 | 0.746 | 0.1957 | No | ||
| 203 | PDK4 | 12624 | 0.650 | 0.1825 | No | ||
| 204 | BCKDHA | 12635 | 0.648 | 0.1825 | No | ||
| 205 | SLC22A4 | 12790 | 0.590 | 0.1758 | No | ||
| 206 | GATM | 12818 | 0.582 | 0.1749 | No | ||
| 207 | NDUFA4 | 13200 | 0.434 | 0.1576 | No | ||
| 208 | POLG2 | 13208 | 0.431 | 0.1576 | No | ||
| 209 | BNIP3 | 13599 | 0.273 | 0.1397 | No | ||
| 210 | SPG7 | 13775 | 0.182 | 0.1318 | No | ||
| 211 | PMAIP1 | 13796 | 0.171 | 0.1310 | No | ||
| 212 | ACADSB | 13976 | 0.092 | 0.1228 | No | ||
| 213 | MOSC2 | 14014 | 0.071 | 0.1211 | No | ||
| 214 | SLC25A22 | 14157 | 0.000 | 0.1145 | No | ||
| 215 | COX6B2 | 14493 | -0.217 | 0.0992 | No | ||
| 216 | ALDH5A1 | 14522 | -0.233 | 0.0981 | No | ||
| 217 | BCKDHB | 14534 | -0.238 | 0.0977 | No | ||
| 218 | CASQ1 | 14822 | -0.430 | 0.0848 | No | ||
| 219 | COX7A1 | 15067 | -0.585 | 0.0739 | No | ||
| 220 | FDX1 | 15071 | -0.586 | 0.0741 | No | ||
| 221 | NDUFA2 | 15080 | -0.594 | 0.0742 | No | ||
| 222 | MRPS21 | 15125 | -0.632 | 0.0725 | No | ||
| 223 | CPS1 | 15245 | -0.707 | 0.0675 | No | ||
| 224 | COQ4 | 15271 | -0.720 | 0.0668 | No | ||
| 225 | HSD3B2 | 15487 | -0.870 | 0.0575 | No | ||
| 226 | ASAH2 | 16170 | -1.388 | 0.0269 | No | ||
| 227 | CKMT1A | 16317 | -1.504 | 0.0212 | No | ||
| 228 | SDHA | 16338 | -1.516 | 0.0213 | No | ||
| 229 | SLC25A27 | 16391 | -1.567 | 0.0199 | No | ||
| 230 | CLN3 | 16411 | -1.584 | 0.0201 | No | ||
| 231 | SURF1 | 16447 | -1.610 | 0.0195 | No | ||
| 232 | CPOX | 16576 | -1.731 | 0.0148 | No | ||
| 233 | ABAT | 16646 | -1.793 | 0.0128 | No | ||
| 234 | TFAM | 16678 | -1.821 | 0.0126 | No | ||
| 235 | OXCT1 | 16713 | -1.850 | 0.0122 | No | ||
| 236 | MSRB2 | 16772 | -1.905 | 0.0108 | No | ||
| 237 | COX11 | 16817 | -1.942 | 0.0101 | No | ||
| 238 | PRDX5 | 16892 | -2.024 | 0.0080 | No | ||
| 239 | DIABLO | 16978 | -2.104 | 0.0055 | No | ||
| 240 | NR3C1 | 17008 | -2.137 | 0.0056 | No | ||
| 241 | NDUFA13 | 17104 | -2.224 | 0.0026 | No | ||
| 242 | MTERF | 17105 | -2.224 | 0.0041 | No | ||
| 243 | TIMM17B | 17123 | -2.238 | 0.0048 | No | ||
| 244 | MTIF2 | 17169 | -2.282 | 0.0043 | No | ||
| 245 | NFS1 | 17182 | -2.290 | 0.0052 | No | ||
| 246 | AMACR | 17234 | -2.330 | 0.0044 | No | ||
| 247 | ACADVL | 17253 | -2.349 | 0.0052 | No | ||
| 248 | ACADM | 17566 | -2.662 | -0.0075 | No | ||
| 249 | BZRAP1 | 17576 | -2.674 | -0.0061 | No | ||
| 250 | RHOT1 | 17585 | -2.679 | -0.0047 | No | ||
| 251 | TOMM34 | 17627 | -2.718 | -0.0048 | No | ||
| 252 | ALDH4A1 | 17697 | -2.780 | -0.0061 | No | ||
| 253 | NIPSNAP1 | 17724 | -2.803 | -0.0055 | No | ||
| 254 | MFN2 | 17850 | -2.947 | -0.0093 | No | ||
| 255 | MYLC2PL | 17853 | -2.948 | -0.0074 | No | ||
| 256 | IDH2 | 17856 | -2.950 | -0.0055 | No | ||
| 257 | PTS | 17871 | -2.965 | -0.0042 | No | ||
| 258 | NDUFS4 | 18111 | -3.233 | -0.0131 | No | ||
| 259 | OGDH | 18246 | -3.378 | -0.0170 | No | ||
| 260 | MPV17 | 18304 | -3.429 | -0.0174 | No | ||
| 261 | SLC25A14 | 18364 | -3.495 | -0.0178 | No | ||
| 262 | MRPL52 | 18366 | -3.498 | -0.0155 | No | ||
| 263 | ACAT1 | 18558 | -3.713 | -0.0218 | No | ||
| 264 | CCDC39 | 18601 | -3.758 | -0.0213 | No | ||
| 265 | BBC3 | 18733 | -3.923 | -0.0247 | No | ||
| 266 | FIS1 | 18905 | -4.153 | -0.0299 | No | ||
| 267 | POLRMT | 18956 | -4.220 | -0.0294 | No | ||
| 268 | SLC25A12 | 19006 | -4.291 | -0.0288 | No | ||
| 269 | SLC9A6 | 19052 | -4.342 | -0.0280 | No | ||
| 270 | RAF1 | 19074 | -4.383 | -0.0260 | No | ||
| 271 | PMPCA | 19176 | -4.544 | -0.0277 | No | ||
| 272 | BPHL | 19210 | -4.593 | -0.0261 | No | ||
| 273 | ACOT9 | 19232 | -4.627 | -0.0240 | No | ||
| 274 | PINK1 | 19279 | -4.705 | -0.0230 | No | ||
| 275 | CLPX | 19385 | -4.869 | -0.0246 | No | ||
| 276 | DGUOK | 19438 | -4.955 | -0.0237 | No | ||
| 277 | BCAT2 | 19588 | -5.168 | -0.0272 | No | ||
| 278 | ACO2 | 19662 | -5.276 | -0.0270 | No | ||
| 279 | PDK1 | 19800 | -5.491 | -0.0297 | No | ||
| 280 | SLC25A11 | 19809 | -5.510 | -0.0264 | No | ||
| 281 | RHOT2 | 20058 | -5.996 | -0.0339 | No | ||
| 282 | GBAS | 20074 | -6.029 | -0.0305 | No | ||
| 283 | TRAK1 | 20477 | -6.919 | -0.0445 | No | ||
| 284 | DBT | 20589 | -7.169 | -0.0449 | No | ||
| 285 | FIBP | 20689 | -7.465 | -0.0445 | No | ||
| 286 | ATPIF1 | 20695 | -7.479 | -0.0397 | No | ||
| 287 | ALAS2 | 20954 | -8.381 | -0.0461 | No | ||
| 288 | POLG | 21020 | -8.661 | -0.0433 | No | ||
| 289 | MCL1 | 21326 | -10.029 | -0.0507 | No | ||
| 290 | PGS1 | 21339 | -10.118 | -0.0445 | No | ||
| 291 | BCL2 | 21371 | -10.300 | -0.0391 | No | ||
| 292 | BCL2L1 | 21524 | -11.498 | -0.0385 | No | ||
| 293 | PDHA1 | 21621 | -12.582 | -0.0346 | No | ||
| 294 | CRY1 | 21920 | -22.141 | -0.0336 | No | ||
| 295 | NAPG | 21923 | -22.494 | -0.0187 | No | ||
| 296 | CASP7 | 21938 | -29.718 | 0.0004 | No |