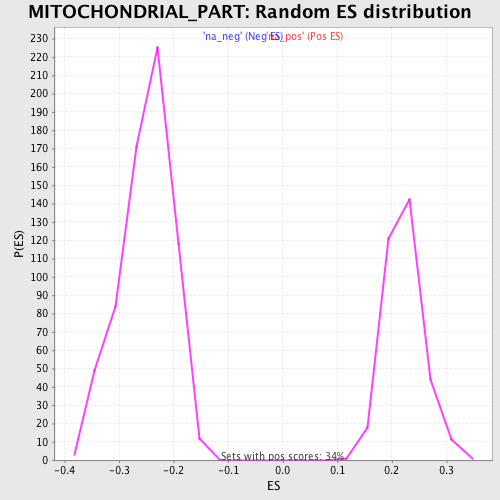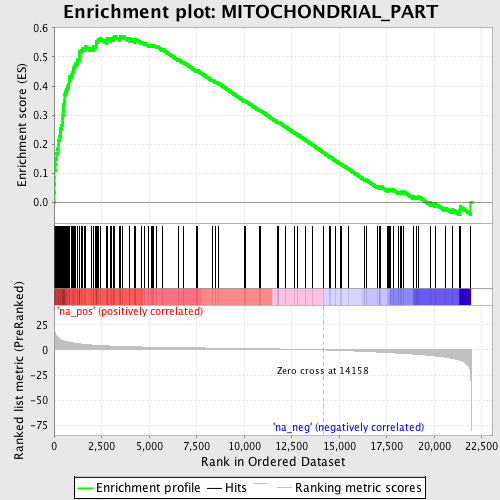
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set01_ATM_minus_versus_ATM_plus |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_pos |
| GeneSet | MITOCHONDRIAL_PART |
| Enrichment Score (ES) | 0.57127464 |
| Normalized Enrichment Score (NES) | 2.574074 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.0 |
| FWER p-Value | 0.0 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | MRPS10 | 4 | 25.608 | 0.0352 | Yes | ||
| 2 | ATP5G1 | 16 | 21.522 | 0.0645 | Yes | ||
| 3 | CYCS | 42 | 17.128 | 0.0871 | Yes | ||
| 4 | TIMM17A | 44 | 16.991 | 0.1105 | Yes | ||
| 5 | PHB2 | 65 | 15.962 | 0.1317 | Yes | ||
| 6 | IMMT | 98 | 14.631 | 0.1505 | Yes | ||
| 7 | PHB | 132 | 13.657 | 0.1678 | Yes | ||
| 8 | TIMM50 | 161 | 12.723 | 0.1842 | Yes | ||
| 9 | VDAC2 | 214 | 11.747 | 0.1980 | Yes | ||
| 10 | GRPEL1 | 220 | 11.637 | 0.2139 | Yes | ||
| 11 | MRPS36 | 263 | 11.174 | 0.2274 | Yes | ||
| 12 | MRPL12 | 316 | 10.627 | 0.2397 | Yes | ||
| 13 | VDAC1 | 339 | 10.409 | 0.2531 | Yes | ||
| 14 | ETFA | 385 | 9.964 | 0.2649 | Yes | ||
| 15 | MRPS11 | 421 | 9.615 | 0.2766 | Yes | ||
| 16 | NDUFV1 | 429 | 9.497 | 0.2894 | Yes | ||
| 17 | NDUFAB1 | 465 | 9.257 | 0.3006 | Yes | ||
| 18 | SDHD | 480 | 9.166 | 0.3126 | Yes | ||
| 19 | TIMM9 | 489 | 9.084 | 0.3248 | Yes | ||
| 20 | VDAC3 | 516 | 8.932 | 0.3360 | Yes | ||
| 21 | MRPL40 | 524 | 8.847 | 0.3479 | Yes | ||
| 22 | TIMM10 | 566 | 8.600 | 0.3579 | Yes | ||
| 23 | MRPS18A | 567 | 8.597 | 0.3698 | Yes | ||
| 24 | SLC25A3 | 603 | 8.417 | 0.3799 | Yes | ||
| 25 | MRPS12 | 666 | 8.123 | 0.3883 | Yes | ||
| 26 | MRPS22 | 705 | 7.954 | 0.3975 | Yes | ||
| 27 | MRPS35 | 746 | 7.786 | 0.4065 | Yes | ||
| 28 | MRPS18C | 783 | 7.653 | 0.4154 | Yes | ||
| 29 | MRPL51 | 811 | 7.526 | 0.4246 | Yes | ||
| 30 | TIMM8B | 832 | 7.464 | 0.4340 | Yes | ||
| 31 | MRPS28 | 930 | 7.107 | 0.4394 | Yes | ||
| 32 | MRPS15 | 953 | 7.020 | 0.4481 | Yes | ||
| 33 | OPA1 | 1003 | 6.867 | 0.4553 | Yes | ||
| 34 | ABCF2 | 1043 | 6.750 | 0.4629 | Yes | ||
| 35 | ATP5J | 1098 | 6.550 | 0.4695 | Yes | ||
| 36 | NDUFS8 | 1146 | 6.442 | 0.4762 | Yes | ||
| 37 | MRPL55 | 1231 | 6.214 | 0.4810 | Yes | ||
| 38 | NDUFA9 | 1233 | 6.204 | 0.4895 | Yes | ||
| 39 | TOMM22 | 1315 | 6.000 | 0.4941 | Yes | ||
| 40 | UQCRC1 | 1327 | 5.967 | 0.5018 | Yes | ||
| 41 | CS | 1336 | 5.959 | 0.5097 | Yes | ||
| 42 | NDUFS3 | 1351 | 5.899 | 0.5172 | Yes | ||
| 43 | ABCB7 | 1428 | 5.759 | 0.5217 | Yes | ||
| 44 | OXA1L | 1476 | 5.629 | 0.5273 | Yes | ||
| 45 | ABCB6 | 1580 | 5.452 | 0.5302 | Yes | ||
| 46 | ATP5F1 | 1653 | 5.316 | 0.5342 | Yes | ||
| 47 | TIMM44 | 1946 | 4.883 | 0.5276 | Yes | ||
| 48 | ABCB8 | 2055 | 4.740 | 0.5292 | Yes | ||
| 49 | ATP5A1 | 2058 | 4.738 | 0.5357 | Yes | ||
| 50 | BAX | 2201 | 4.543 | 0.5354 | Yes | ||
| 51 | ATP5O | 2209 | 4.538 | 0.5414 | Yes | ||
| 52 | MRPL23 | 2213 | 4.530 | 0.5475 | Yes | ||
| 53 | SLC25A1 | 2216 | 4.528 | 0.5537 | Yes | ||
| 54 | ATP5E | 2306 | 4.413 | 0.5557 | Yes | ||
| 55 | MRPL32 | 2334 | 4.378 | 0.5605 | Yes | ||
| 56 | TIMM23 | 2418 | 4.296 | 0.5627 | Yes | ||
| 57 | ATP5G3 | 2767 | 3.974 | 0.5522 | Yes | ||
| 58 | ACN9 | 2788 | 3.961 | 0.5568 | Yes | ||
| 59 | ATP5D | 2793 | 3.957 | 0.5621 | Yes | ||
| 60 | TIMM13 | 2968 | 3.821 | 0.5594 | Yes | ||
| 61 | SLC25A15 | 3014 | 3.790 | 0.5626 | Yes | ||
| 62 | SUPV3L1 | 3135 | 3.706 | 0.5622 | Yes | ||
| 63 | MSTO1 | 3139 | 3.704 | 0.5672 | Yes | ||
| 64 | PITRM1 | 3162 | 3.688 | 0.5713 | Yes | ||
| 65 | BCS1L | 3457 | 3.457 | 0.5626 | No | ||
| 66 | HTRA2 | 3480 | 3.446 | 0.5663 | No | ||
| 67 | NDUFB6 | 3505 | 3.432 | 0.5700 | No | ||
| 68 | ATP5G2 | 3603 | 3.359 | 0.5702 | No | ||
| 69 | ATP5C1 | 3949 | 3.174 | 0.5588 | No | ||
| 70 | ATP5B | 3951 | 3.174 | 0.5631 | No | ||
| 71 | MRPS16 | 4239 | 3.023 | 0.5541 | No | ||
| 72 | UQCRB | 4258 | 3.014 | 0.5575 | No | ||
| 73 | MTX2 | 4291 | 3.000 | 0.5602 | No | ||
| 74 | NNT | 4589 | 2.880 | 0.5505 | No | ||
| 75 | DNAJA3 | 4764 | 2.809 | 0.5464 | No | ||
| 76 | MRPL41 | 4974 | 2.731 | 0.5406 | No | ||
| 77 | COX15 | 5112 | 2.683 | 0.5381 | No | ||
| 78 | NDUFS7 | 5194 | 2.655 | 0.5380 | No | ||
| 79 | NDUFA1 | 5233 | 2.644 | 0.5400 | No | ||
| 80 | ETFB | 5412 | 2.587 | 0.5354 | No | ||
| 81 | NDUFA6 | 5703 | 2.494 | 0.5255 | No | ||
| 82 | UCP3 | 6536 | 2.242 | 0.4905 | No | ||
| 83 | RAB11FIP5 | 6791 | 2.174 | 0.4819 | No | ||
| 84 | BCKDK | 7507 | 1.985 | 0.4518 | No | ||
| 85 | PPOX | 7570 | 1.970 | 0.4517 | No | ||
| 86 | NDUFS2 | 8317 | 1.784 | 0.4200 | No | ||
| 87 | MAOB | 8508 | 1.742 | 0.4137 | No | ||
| 88 | UQCRH | 8656 | 1.702 | 0.4093 | No | ||
| 89 | HCCS | 10013 | 1.379 | 0.3491 | No | ||
| 90 | TFB2M | 10069 | 1.366 | 0.3484 | No | ||
| 91 | HSD3B1 | 10809 | 1.185 | 0.3162 | No | ||
| 92 | CENTA2 | 10854 | 1.174 | 0.3158 | No | ||
| 93 | HADHB | 11770 | 0.914 | 0.2751 | No | ||
| 94 | MRPL10 | 11785 | 0.910 | 0.2757 | No | ||
| 95 | NDUFS1 | 11834 | 0.895 | 0.2748 | No | ||
| 96 | MRPS24 | 12152 | 0.797 | 0.2613 | No | ||
| 97 | BCKDHA | 12635 | 0.648 | 0.2401 | No | ||
| 98 | GATM | 12818 | 0.582 | 0.2326 | No | ||
| 99 | POLG2 | 13208 | 0.431 | 0.2154 | No | ||
| 100 | BNIP3 | 13599 | 0.273 | 0.1979 | No | ||
| 101 | SLC25A22 | 14157 | 0.000 | 0.1723 | No | ||
| 102 | COX6B2 | 14493 | -0.217 | 0.1573 | No | ||
| 103 | BCKDHB | 14534 | -0.238 | 0.1558 | No | ||
| 104 | CASQ1 | 14822 | -0.430 | 0.1432 | No | ||
| 105 | NDUFA2 | 15080 | -0.594 | 0.1323 | No | ||
| 106 | MRPS21 | 15125 | -0.632 | 0.1311 | No | ||
| 107 | HSD3B2 | 15487 | -0.870 | 0.1158 | No | ||
| 108 | SDHA | 16338 | -1.516 | 0.0789 | No | ||
| 109 | SURF1 | 16447 | -1.610 | 0.0762 | No | ||
| 110 | NR3C1 | 17008 | -2.137 | 0.0535 | No | ||
| 111 | NDUFA13 | 17104 | -2.224 | 0.0522 | No | ||
| 112 | TIMM17B | 17123 | -2.238 | 0.0545 | No | ||
| 113 | NFS1 | 17182 | -2.290 | 0.0550 | No | ||
| 114 | ACADM | 17566 | -2.662 | 0.0411 | No | ||
| 115 | RHOT1 | 17585 | -2.679 | 0.0440 | No | ||
| 116 | TOMM34 | 17627 | -2.718 | 0.0459 | No | ||
| 117 | ALDH4A1 | 17697 | -2.780 | 0.0466 | No | ||
| 118 | MFN2 | 17850 | -2.947 | 0.0437 | No | ||
| 119 | NDUFS4 | 18111 | -3.233 | 0.0362 | No | ||
| 120 | OGDH | 18246 | -3.378 | 0.0347 | No | ||
| 121 | MPV17 | 18304 | -3.429 | 0.0369 | No | ||
| 122 | MRPL52 | 18366 | -3.498 | 0.0389 | No | ||
| 123 | FIS1 | 18905 | -4.153 | 0.0200 | No | ||
| 124 | RAF1 | 19074 | -4.383 | 0.0184 | No | ||
| 125 | PMPCA | 19176 | -4.544 | 0.0200 | No | ||
| 126 | SLC25A11 | 19809 | -5.510 | -0.0013 | No | ||
| 127 | RHOT2 | 20058 | -5.996 | -0.0044 | No | ||
| 128 | DBT | 20589 | -7.169 | -0.0188 | No | ||
| 129 | ALAS2 | 20954 | -8.381 | -0.0239 | No | ||
| 130 | MCL1 | 21326 | -10.029 | -0.0270 | No | ||
| 131 | BCL2 | 21371 | -10.300 | -0.0148 | No | ||
| 132 | CASP7 | 21938 | -29.718 | 0.0004 | No |