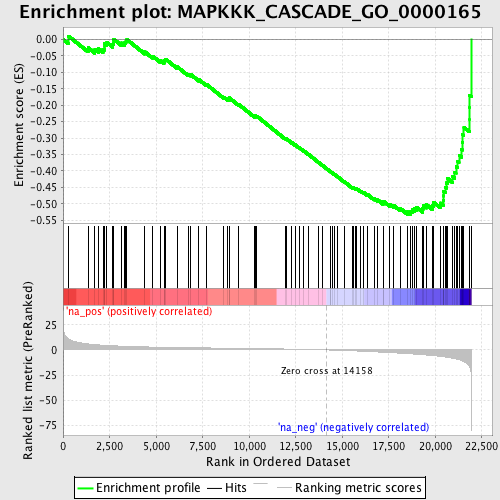
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set01_ATM_minus_versus_ATM_plus |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_neg |
| GeneSet | MAPKKK_CASCADE_GO_0000165 |
| Enrichment Score (ES) | -0.53274095 |
| Normalized Enrichment Score (NES) | -2.0443757 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.012704406 |
| FWER p-Value | 0.092 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | DUSP9 | 291 | 10.801 | 0.0104 | No | ||
| 2 | DUSP22 | 1346 | 5.915 | -0.0249 | No | ||
| 3 | C5AR1 | 1709 | 5.233 | -0.0300 | No | ||
| 4 | MAPK8IP1 | 1902 | 4.938 | -0.0279 | No | ||
| 5 | SOD1 | 2146 | 4.618 | -0.0289 | No | ||
| 6 | TGFA | 2218 | 4.525 | -0.0223 | No | ||
| 7 | ADRA2A | 2222 | 4.513 | -0.0125 | No | ||
| 8 | NRTN | 2352 | 4.360 | -0.0088 | No | ||
| 9 | PROK2 | 2669 | 4.044 | -0.0144 | No | ||
| 10 | EDG5 | 2714 | 4.003 | -0.0077 | No | ||
| 11 | MAPKAPK2 | 2720 | 3.998 | 0.0009 | No | ||
| 12 | SCG2 | 3146 | 3.697 | -0.0105 | No | ||
| 13 | ADRA2C | 3314 | 3.569 | -0.0103 | No | ||
| 14 | FPR1 | 3376 | 3.516 | -0.0054 | No | ||
| 15 | MAP3K6 | 3427 | 3.476 | -0.0000 | No | ||
| 16 | GHRL | 4394 | 2.955 | -0.0378 | No | ||
| 17 | PTPLAD1 | 4805 | 2.791 | -0.0504 | No | ||
| 18 | MAP3K9 | 5225 | 2.645 | -0.0638 | No | ||
| 19 | MAPK10 | 5422 | 2.586 | -0.0671 | No | ||
| 20 | ADORA2B | 5461 | 2.570 | -0.0632 | No | ||
| 21 | FGFR3 | 5506 | 2.555 | -0.0596 | No | ||
| 22 | ADRB3 | 6121 | 2.360 | -0.0825 | No | ||
| 23 | FGF2 | 6738 | 2.186 | -0.1059 | No | ||
| 24 | FGFR1 | 6855 | 2.152 | -0.1065 | No | ||
| 25 | MAPK8IP2 | 7294 | 2.039 | -0.1220 | No | ||
| 26 | MDFI | 7712 | 1.927 | -0.1369 | No | ||
| 27 | MAP3K2 | 8636 | 1.709 | -0.1754 | No | ||
| 28 | DUSP8 | 8849 | 1.661 | -0.1814 | No | ||
| 29 | CD24 | 8918 | 1.644 | -0.1810 | No | ||
| 30 | TPD52L1 | 8920 | 1.643 | -0.1774 | No | ||
| 31 | MAP3K4 | 9437 | 1.520 | -0.1977 | No | ||
| 32 | EDA2R | 10305 | 1.311 | -0.2345 | No | ||
| 33 | CHRNA7 | 10311 | 1.308 | -0.2318 | No | ||
| 34 | GNG3 | 10401 | 1.286 | -0.2331 | No | ||
| 35 | GRM4 | 11922 | 0.864 | -0.3007 | No | ||
| 36 | DUSP10 | 11980 | 0.847 | -0.3015 | No | ||
| 37 | EGF | 12295 | 0.756 | -0.3142 | No | ||
| 38 | TDGF1 | 12467 | 0.703 | -0.3205 | No | ||
| 39 | NF1 | 12700 | 0.627 | -0.3297 | No | ||
| 40 | CD81 | 12940 | 0.537 | -0.3395 | No | ||
| 41 | RGS4 | 13168 | 0.450 | -0.3489 | No | ||
| 42 | GADD45G | 13718 | 0.218 | -0.3735 | No | ||
| 43 | AMBP | 13939 | 0.112 | -0.3833 | No | ||
| 44 | GPS2 | 14368 | -0.134 | -0.4026 | No | ||
| 45 | CARTPT | 14372 | -0.135 | -0.4025 | No | ||
| 46 | ATP6AP2 | 14483 | -0.212 | -0.4070 | No | ||
| 47 | PLCE1 | 14566 | -0.256 | -0.4102 | No | ||
| 48 | DUSP4 | 14744 | -0.376 | -0.4175 | No | ||
| 49 | PIK3CB | 15122 | -0.630 | -0.4334 | No | ||
| 50 | TRAF7 | 15556 | -0.921 | -0.4512 | No | ||
| 51 | GPS1 | 15608 | -0.951 | -0.4514 | No | ||
| 52 | MBIP | 15706 | -1.018 | -0.4536 | No | ||
| 53 | MAP4K3 | 15752 | -1.048 | -0.4534 | No | ||
| 54 | ADRB2 | 15984 | -1.226 | -0.4613 | No | ||
| 55 | CAMKK2 | 16123 | -1.344 | -0.4646 | No | ||
| 56 | DAXX | 16329 | -1.510 | -0.4707 | No | ||
| 57 | SHC1 | 16723 | -1.857 | -0.4846 | No | ||
| 58 | TNIK | 16901 | -2.033 | -0.4883 | No | ||
| 59 | MAP3K7IP1 | 17192 | -2.295 | -0.4965 | No | ||
| 60 | MAPK9 | 17214 | -2.310 | -0.4924 | No | ||
| 61 | SPRED1 | 17558 | -2.657 | -0.5023 | No | ||
| 62 | PAK1 | 17746 | -2.826 | -0.5046 | No | ||
| 63 | RGS3 | 18102 | -3.221 | -0.5138 | No | ||
| 64 | DBNL | 18517 | -3.675 | -0.5247 | Yes | ||
| 65 | DUSP6 | 18680 | -3.854 | -0.5236 | Yes | ||
| 66 | MAP3K10 | 18760 | -3.957 | -0.5186 | Yes | ||
| 67 | CXCR4 | 18871 | -4.113 | -0.5146 | Yes | ||
| 68 | HIPK2 | 18997 | -4.265 | -0.5110 | Yes | ||
| 69 | MAP3K12 | 19326 | -4.778 | -0.5155 | Yes | ||
| 70 | MAP3K11 | 19341 | -4.796 | -0.5056 | Yes | ||
| 71 | SH2D3C | 19505 | -5.042 | -0.5020 | Yes | ||
| 72 | MDFIC | 19856 | -5.585 | -0.5058 | Yes | ||
| 73 | LAX1 | 19906 | -5.674 | -0.4956 | Yes | ||
| 74 | MAP2K7 | 20261 | -6.417 | -0.4977 | Yes | ||
| 75 | MAP3K3 | 20422 | -6.791 | -0.4902 | Yes | ||
| 76 | MAP3K13 | 20449 | -6.853 | -0.4763 | Yes | ||
| 77 | PKN1 | 20455 | -6.883 | -0.4615 | Yes | ||
| 78 | CRKL | 20555 | -7.087 | -0.4504 | Yes | ||
| 79 | SPRED2 | 20599 | -7.209 | -0.4366 | Yes | ||
| 80 | MINK1 | 20643 | -7.336 | -0.4225 | Yes | ||
| 81 | DUSP16 | 20921 | -8.252 | -0.4171 | Yes | ||
| 82 | TAOK3 | 21049 | -8.758 | -0.4037 | Yes | ||
| 83 | DUSP2 | 21113 | -8.980 | -0.3869 | Yes | ||
| 84 | MAPK8 | 21208 | -9.408 | -0.3705 | Yes | ||
| 85 | MADD | 21274 | -9.747 | -0.3521 | Yes | ||
| 86 | GADD45B | 21406 | -10.572 | -0.3350 | Yes | ||
| 87 | MAP4K5 | 21442 | -10.831 | -0.3128 | Yes | ||
| 88 | MAP2K4 | 21466 | -10.991 | -0.2898 | Yes | ||
| 89 | CCM2 | 21538 | -11.589 | -0.2676 | Yes | ||
| 90 | TAOK2 | 21830 | -16.810 | -0.2440 | Yes | ||
| 91 | MAPK8IP3 | 21835 | -16.890 | -0.2072 | Yes | ||
| 92 | MAP4K2 | 21842 | -17.169 | -0.1698 | Yes | ||
| 93 | FGF13 | 21947 | -79.619 | -0.0000 | Yes |