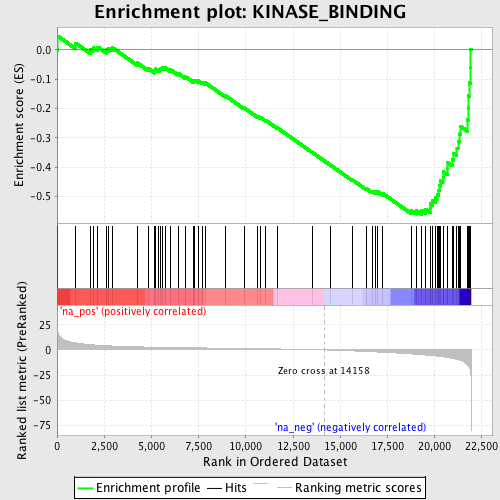
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set01_ATM_minus_versus_ATM_plus |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_neg |
| GeneSet | KINASE_BINDING |
| Enrichment Score (ES) | -0.5608785 |
| Normalized Enrichment Score (NES) | -2.0087507 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.015501255 |
| FWER p-Value | 0.139 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | HINT1 | 38 | 17.597 | 0.0467 | No | ||
| 2 | CDK5RAP2 | 980 | 6.934 | 0.0228 | No | ||
| 3 | CDKN2C | 1747 | 5.171 | 0.0020 | No | ||
| 4 | MAPK8IP1 | 1902 | 4.938 | 0.0086 | No | ||
| 5 | CDKN2A | 2159 | 4.589 | 0.0095 | No | ||
| 6 | PRDX3 | 2612 | 4.097 | 0.0002 | No | ||
| 7 | ZC3HC1 | 2723 | 3.997 | 0.0061 | No | ||
| 8 | FAF1 | 2945 | 3.834 | 0.0066 | No | ||
| 9 | AKAP3 | 4247 | 3.020 | -0.0445 | No | ||
| 10 | AVPR1A | 4847 | 2.777 | -0.0643 | No | ||
| 11 | ADAM9 | 5171 | 2.662 | -0.0717 | No | ||
| 12 | TOP2A | 5189 | 2.657 | -0.0651 | No | ||
| 13 | AKAP12 | 5361 | 2.605 | -0.0658 | No | ||
| 14 | YWHAG | 5501 | 2.556 | -0.0651 | No | ||
| 15 | INSR | 5561 | 2.539 | -0.0608 | No | ||
| 16 | TPRKB | 5722 | 2.488 | -0.0613 | No | ||
| 17 | AKAP7 | 6008 | 2.392 | -0.0677 | No | ||
| 18 | ADAM10 | 6405 | 2.277 | -0.0795 | No | ||
| 19 | MOBKL1A | 6817 | 2.166 | -0.0924 | No | ||
| 20 | TRIB3 | 7235 | 2.055 | -0.1058 | No | ||
| 21 | MAPK8IP2 | 7294 | 2.039 | -0.1028 | No | ||
| 22 | LATS1 | 7482 | 1.992 | -0.1058 | No | ||
| 23 | AKAP4 | 7701 | 1.929 | -0.1105 | No | ||
| 24 | IRAK1 | 7859 | 1.892 | -0.1125 | No | ||
| 25 | CD24 | 8918 | 1.644 | -0.1563 | No | ||
| 26 | AKAP5 | 9904 | 1.409 | -0.1974 | No | ||
| 27 | SPDYA | 10604 | 1.237 | -0.2260 | No | ||
| 28 | PTPRR | 10798 | 1.187 | -0.2315 | No | ||
| 29 | BCAR1 | 11044 | 1.123 | -0.2396 | No | ||
| 30 | AVPR1B | 11661 | 0.948 | -0.2652 | No | ||
| 31 | AKAP11 | 13528 | 0.305 | -0.3496 | No | ||
| 32 | CDKN2B | 14499 | -0.220 | -0.3933 | No | ||
| 33 | IRS1 | 15669 | -0.991 | -0.4440 | No | ||
| 34 | GRK5 | 16384 | -1.558 | -0.4723 | No | ||
| 35 | FIZ1 | 16703 | -1.842 | -0.4818 | No | ||
| 36 | RPS6KA4 | 16863 | -1.997 | -0.4836 | No | ||
| 37 | PDLIM5 | 16985 | -2.109 | -0.4833 | No | ||
| 38 | PICK1 | 17237 | -2.332 | -0.4884 | No | ||
| 39 | RB1 | 18763 | -3.958 | -0.5471 | No | ||
| 40 | TRIP6 | 19029 | -4.323 | -0.5474 | No | ||
| 41 | MAP3K12 | 19326 | -4.778 | -0.5477 | Yes | ||
| 42 | PTPRC | 19517 | -5.062 | -0.5425 | Yes | ||
| 43 | TOP2B | 19769 | -5.434 | -0.5390 | Yes | ||
| 44 | APC | 19779 | -5.445 | -0.5244 | Yes | ||
| 45 | LAX1 | 19906 | -5.674 | -0.5145 | Yes | ||
| 46 | CAMK2N1 | 20039 | -5.959 | -0.5041 | Yes | ||
| 47 | KIF13B | 20132 | -6.134 | -0.4915 | Yes | ||
| 48 | BCL10 | 20231 | -6.352 | -0.4784 | Yes | ||
| 49 | MAP2K7 | 20261 | -6.417 | -0.4621 | Yes | ||
| 50 | ITGB2 | 20306 | -6.502 | -0.4462 | Yes | ||
| 51 | MAP3K13 | 20449 | -6.853 | -0.4338 | Yes | ||
| 52 | UBQLN1 | 20466 | -6.903 | -0.4155 | Yes | ||
| 53 | RAD9A | 20699 | -7.495 | -0.4055 | Yes | ||
| 54 | DLG1 | 20702 | -7.506 | -0.3849 | Yes | ||
| 55 | CEP250 | 20937 | -8.326 | -0.3727 | Yes | ||
| 56 | RELA | 21022 | -8.671 | -0.3527 | Yes | ||
| 57 | CD4 | 21188 | -9.300 | -0.3346 | Yes | ||
| 58 | LLGL1 | 21248 | -9.615 | -0.3108 | Yes | ||
| 59 | SQSTM1 | 21318 | -9.993 | -0.2864 | Yes | ||
| 60 | SOCS1 | 21356 | -10.225 | -0.2600 | Yes | ||
| 61 | SIT1 | 21731 | -14.291 | -0.2377 | Yes | ||
| 62 | LCK | 21778 | -15.166 | -0.1981 | Yes | ||
| 63 | IGF1R | 21784 | -15.295 | -0.1562 | Yes | ||
| 64 | MAPK8IP3 | 21835 | -16.890 | -0.1119 | Yes | ||
| 65 | CDKN2D | 21895 | -19.642 | -0.0605 | Yes | ||
| 66 | CCND3 | 21924 | -22.833 | 0.0011 | Yes |
